8.5: Structure and Organization of DNA in Bacteria
- Page ID
- 16461
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Sexual reproduction allows compatible genders (think male and female) to share genes, a strategy that increases species diversity. It turns out that bacteria and other single celled organisms can also share genes… and spread diversity. We will close this chapter with a look at sex (E. coli style!), and gene-mapping experiments showing linearly arranged genes on a circular bacterial DNA molecule (the bacterial ‘chromosome’).
E. coli sex begins when F+ and F- cells meet. These cells are “opposite” mating types that can share DNA during conjugation. F+ cells contain the F plasmid, a small circular DNA molecule that is separate from the E. coli chromosome. The F (fertility) plasmid has genes that encode sex pili on F+, as well as factors needed to form a mating bridge, or conjugation tube. The behavior of the F plasmid during conjugation is shown below.
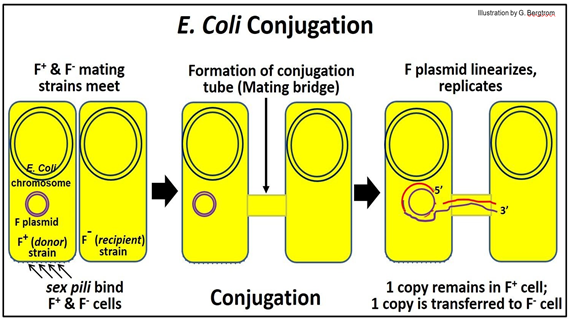
When an F+ (donor) cell encounters an F- (recipient) cell, sex pili on the donor cell initiate recognition. Next, a conjugation tube forms, linking the cytoplasms of the two cells. After nicking one strand of the F plasmid DNA, the nicked begins to roll into the conjugation tube and into the recipient (F-) cell. The DNA strand entering the recipient cell replicates, as does the intact circle remaining in the donor cell (replicating DNA is shown in red in the illustration).
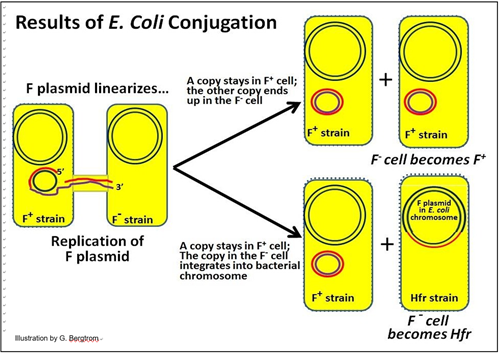
E. coli conjugation can have different outcomes:
1. One outcome is that one of two semi-conservatively replicated F plasmids remains in the donor cell and another is now in the recipient cell. In this case, the recipient cell becomes a new F+ donor cell!
2. The other outcome is integration of the F plasmid into recipient cell chromosomal DNA. Insertion is typically at specific sites in the DNA where there is sufficient sequence similarity between the plasmid and chromosomal DNA to allow insertion by recombination. The result is that the recipient cell becomes an Hfr (High-frequency recombination) cell. This cell will produce Hfr strain progeny cells.
These two possible results of conjugation in E. coli are illustrated below.
Hfr cells readily express their integrated F plasmid genes, and like F+ cells, develop sex pili and form a conjugation tube with an F- cell. One strand of the bacterial chromosomal DNA will be nicked at the original insertion site of the F plasmid.
The next events parallel the replicative transfer of an F plasmid during F+/F- conjugation, except that only part of the Hfr donor chromosomal DNA is transferred, as shown below.
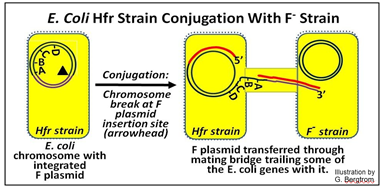
In this illustration, the F plasmid has inserted ‘in front of’ an A gene so that when it enters the conjugation tube, it brings along several E. coli chromosomal genes Because of the size of the bacterial chromosome, only a few bacterial genes enter the recipient gene before transfer is aborted. But in the brief time of DNA transfer, at least some genes did get in to the recipient F- strain where they can be expressed. Here is an outline of an experiment that allowed mapping bacterial genes on a circular DNA chromosome:
1. Hfr cells containing functional A, B, C, D genes were mated with recipient cells containing mutants of either the A, the B, the C or the D gene.
2. Conjugation was mechanically disrupted at different times after the formation of a conjugation tube.
3. Recipient cells from each of the disrupted conjugations were then grown in culture and analyzed for specific gene function.
In this hypothetical example, the results were that a recipient cell with a mutant A gene acquired a wild type A gene (and therefore A-gene function) after a short time before conjugation disruption. Progressively longer times of conjugation (measured in separate experiments) were required to transfer gens B, C and D (respectively) to the recipient cell. Thus the order of these genes on the bacterial chromosome was
-A-B-C-D-
The timing of conjugation that led to F- mutants acquiring a functional gene from the Hfr strain was so refined that not only could the gene locus be determined, but even the size ) length) of the genes! Thus, the time to transfer a complete gene to an F- cell reflects the size (length) of the gene. The other important conclusion is that genes are arranged linearly on bacterial DNA.
Recall that genes already mapped along the length of eukaryotic chromosomes implied a linear order of the genes. However, little was known about eukaryotic chromosome structure at the time, and the role of DNA as the ‘stuff of genes’ was not appreciated. These bacterial mating experiments demonstrated for the first time that genes are linearly arranged not just along a chromosome, but also along the DNA molecule.
Over time, many bacterial genes were mapped all along the E. coli chromosome by isolating many different Hfr strains in which an F plasmid had inserted into different sites around the DNA circle. These Hfr strains were mated to F- bacteria, each with mutations in one or another known bacterial gene. As in the original ‘ABCD’ experiment, the order of many genes was determined, and even shown to be linked at a greater or lesser distance to those ABCD genes and each other.
The map that results from such a study is diagrammed below.
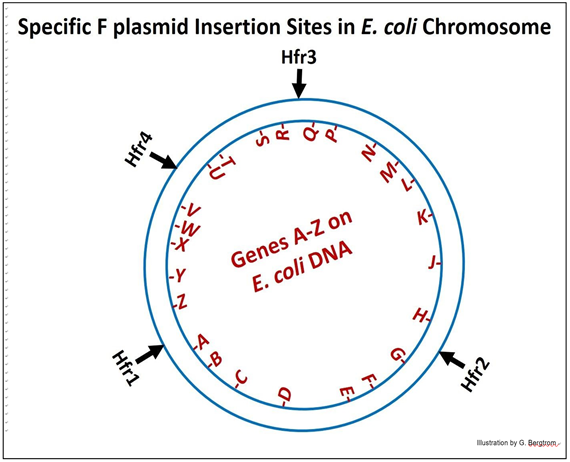
Using the different Hfr strains (numbered in the diagram) in conjugation experiments, it was shown that in fact, the different Hfr cells transferred different genes into the recipient cells in the order implied by the diagram. What’s more, when the experiment was done with Hfr4 (in this generic diagram); the order of genes transferred after longer times of conjugation was found to be:
V-W-X-Y-Z-A-B….
The obvious conclusion from experiments like these was that the E. coli DNA molecule (its ‘chromosome’) was a closed circle! We will see visual evidence of circular E. coli chromosomes in the next chapter, with some discussion of how this evidence informed our understanding of DNA replication.


