5: Chromatin Immunoprecipitation
- Page ID
- 185750
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Summary
Chromatin immunoprecipitation is used to analyze protein-DNA binding interactions.
Also known as:
ChIP
Variants: ChIP-Seq, ChIP-qPCR
Samples needed
ChIP is used to determine whether a specific protein binds to DNA (either generally or at a sequence of interest), so the cells of interest are needed.
Method
Chromatin immunoprecipitation is used to analyze protein-DNA interactions in the cell. The first step in this method is to add a crosslinking agent (e.g., formaldehyde), which covalently binds proteins to DNA, preserving the natural protein-DNA interactions. The cells are lysed to release proteins, DNA, and protein-DNA complexes. Then, the chromatin is fragmented into smaller pieces, typically using sonication. After fragmentation, the lysates are incubated with beads bound to an antibody against the protein of interest. The antibody binds the protein of interest and, consequently, any DNA bound to it.
Next, the beads are washed to get rid of nonspecific binding, and the DNA is eluted from the beads. The crosslinks are reversed to isolate DNA from protein, and the DNA is purified. After purification of the DNA, there are several analysis methods that can be used. If the goal is to quantify the presence of a specific gene/region of DNA, qPCR can be used. (PCR can sometimes be used, but it is less common.) If the goal is to determine several possible genomic sequences present, sequencing methods can be used.
Controls
An input sample should always be used as a control, which is a sample that has not been incubated with the antibody against the protein of interest. The IP samples, which were incubated with the antibody against the protein of interest, are typically normalized to the input samples. Other controls can also be included. For example, an IgG control, where the sample is incubated with a nonspecific antibody, confirms the specificity of the protein-antibody interaction. Positive and negative locus controls use primers that amplify a locus where the protein of interest is either known to bind or known not to bind. These locus controls confirm that the experiment is working correctly and that the protein-DNA interaction is specific.
Interpretation
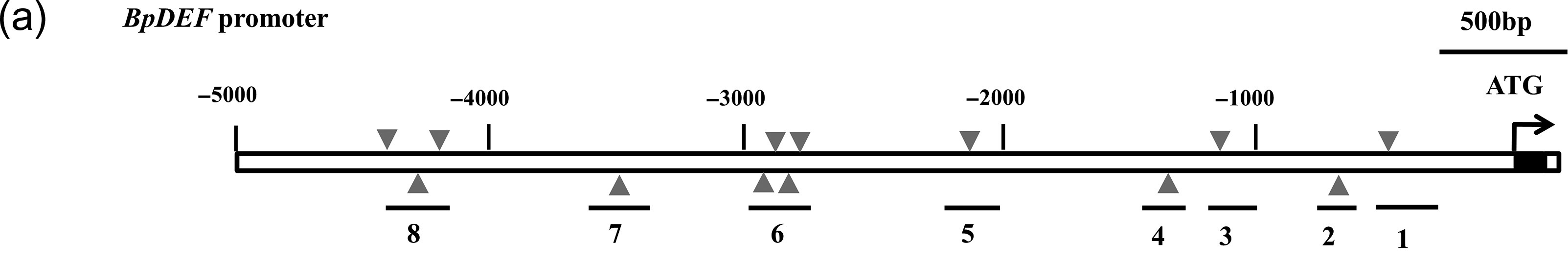

Figure 1. Chromatin immunoprecipitation enrichment of protein BpAP1 at the promoter of the BpDEF gene using qPCR. Relevant section of caption for published figure reads: “ (a) Promoter structure of BpDEF. Gray triangle represents GArG box motif, and DNA fragments near GArG box used for ChIP-PCR are shown. (c) ChIP fold enrichment of DNA fragments around the CArG boxes by ChIP-qPCR.” “Figure 7” by Wang al.[1] [Image description]
BpAP1 is a protein in birch trees that can cause early flowering. In Wang et al., the authors wanted to determine the regulatory mechanism of the BpAP1 protein. To do so, they tested whether BpAP1 was enriched at the promoter of the BpDEF gene, which can promote flowering in birch trees. They tagged the BpAP1 protein with GFP and used an anti-GFP antibody to separate the BpAP1-DNA complexes.
Panel a shows a map of the BpDEF promoter, and each numbered black line represents a region the researchers tested for BpAP1 binding. At each numbered region, they used qPCR to quantify the amount of that sequence of DNA present in their IP samples. The gray bars in the graph depict the fold enrichment of BpAP1-GFP at each of the 8 regions of the BpDEF promoter. The black bars depict the IgG control. Note that the gray bars represent the IP samples over the Input samples for each of the 8 regions, and they are normalized to the IgG control. Based on the results from this experiment, the researchers can conclude that BpAP1 is enriched at (binds to) regions 1, 4, 5, and 6 of the BpDEF promoter.
Image Descriptions
Figure 1 image description:
Panel a: A map of the BpDEF gene promoter, which consists of 5000 base pairs upstream of the start codon. Gray triangles mark GArG box motifs, of which there are 13 total in the promoter. 8 numbered black lines (numbered from 3’ to 5’) mark regions that the researchers amplified using qPCR, and each region contains a GArG motif.
Panel b: A bar graph depicting ChIP fold enrichment in the anti-IgG and anti-GFP samples. The anti-GFP samples are normalized to the anti-IgG samples. The anti-GFP samples have the following enrichment of BpAP1-GFP at each region:
1: 3.3
2: 1.1
3: 1.2
4: 1.9
5: 1.6
6: 1.7
7: 1.3
8: 1.1
BpAP1 is significantly enriched at regions 1, 4, 5, and 6.↵
Thumbnail
"Chip sequencing2" ↗ by Chris Taplin is licensed under CC BY 3.0 ↗.
Description: A simple representation of small fragments of DNA bound by multiple different proteins, and one of those proteins bound by an antibody.
Author
Sara Crippen, Tufts University
-
Wang, S. H. Huang, R. Han, J. Chen, J. Jiang, H. Li, G. Liu, and S. Chen. 2019. BpAP1 directly regulates BpDEF to promote male inflorescence formation in Betula platyphylla x B. pendula. Tree Physiology 39: 1046-1060.↵

