6: Coimmunoprecipitation
- Page ID
- 135770
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Summary
Coimmunoprecipitation is used to determine if two proteins bind to one another in a sample.
Also known as:
Co-IP
Variant: Pull-down. Like a co-IP, a pull-down experiment determines if two proteins bind to one another, but unlike a co-IP, pull-downs do not involve an antibody-antigen interaction. Instead, the first protein of interest is bound to a column or precipitated (“pulled down”) through some other affinity interaction, like 6x His tag with nickel or cobalt ions, or biotin with streptavidin.
However, in practice, the term "pull down" or "pull-down" is often used interchangably with immunoprecipitation.
Samples needed
Usually co-IPs are used to determine if two proteins bind to one another in cells, so the cells of interest are needed.
Method
Coimmunoprecipitation allows a researcher to determine whether two proteins of interest (A and B) bind to one another in a cell lysate or other sample. First, an antibody that is specific to protein A is attached to beads that can be separated from a solution by centrifugation, magnet, or some other method. Then cells of interest are lysed to release the proteins. The lysate is incubated with the anti-A beads as well as control beads that have been attached to an antibody that does not bind protein A. During the incubation, the anti-A antibody binds protein A. Importantly, any other proteins that are also bound to protein A will also be bound to the beads through protein A. The exact buffer used for this incubation and the subsequent washes is important, because the buffer composition will determine what protein-protein interactions are maintained or broken.
Then the beads are washed to remove any proteins that are binding non-specifically, and finally the proteins still bound to the beads are eluted. This leaves the researcher with a solution of protein A and any other proteins that are bound to protein A. Since the question is whether protein A binds protein B, the sample is then subjected to western blot to detect protein B.
Controls
Researchers should always show whole lysate control, or a sample that has not been subjected to immunoprecipitation. This shows the reader how much of the protein of interest is present in the sample to begin with. Also, researchers should show a control where the pull-down is performed with an antibody that does not bind to the protein of interest.
Interpretation
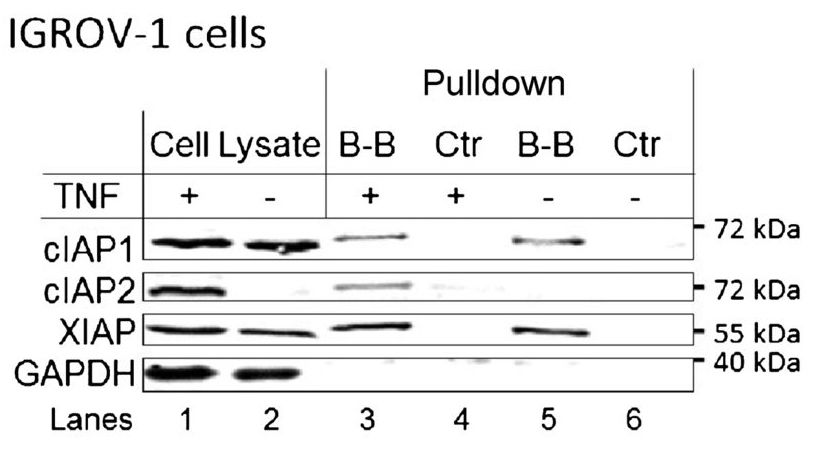
Strictly speaking, one could argue that this is not an immunoprecipitation experiment, because there is no antibody involved. However, the protocol and reasoning are similar, and it is a form of pull-down assay. In this case, the researchers are asking whether the proteins cIAP1, cIAP2, and XIAP bind to the molecule birinapant. A molecule called biotin has been attached to the birinapant. Biotin binds very strongly to another molecule, avidin, which is attached to the magnetic beads. Therefore, the connections are: magnetic beads-avidin-biotin-birinapant-protein that binds birinapant.
Lanes 1 and 2 are the cell lysates before the pull-down is performed. This shows that cIAP1 and XIAP are expressed in the cells regardless of whether they are treated with TNF. cIAP2 is only detectable if the cells are treated with TNF. GAPDH serves as the loading control. Lanes 3 and 4 are the pull-down of lysates from cells treated with TNF; lanes 5 and 6 are the same but without TNF treatment. The control lysates have been treated with beads, but not with biotinylated birinapant. Therefore, as expected, they show no proteins eluted (lanes 4 and 6). Lane 3 shows three bands: one each for cIAP1, cIAP2, and XIAP. This indicates that in the cell lysates, those proteins bind the birinapant molecule. Lane 5 shows the same thing, but since the cells were not treated with TNF, there is no cIAP2 present.
Image Descriptions
Figure 1 image description:
A Western blot. Results are shown in table below.
| Sample | Cell lysate | Cell lysate | B-B pulldown | Ctrl pulldown | B-B pulldown | Ctrl pulldown |
|---|---|---|---|---|---|---|
| TNF treatment? | + | – | + | + | – | – |
| cIAP1 | Band | Band | Band | Band | ||
| cIAP2 | Band | Band | ||||
| XIAP | Band | Band | Band | Band | ||
| GAPDH | Band | Band |
Thumbnail
"Antibody IgG1 surface.png" ↗ by Tokenzero is licensed under CC BY-SA 4.0 ↗.
Description: The structure of an antibody of subclass IgG1. Two light chains (green and pink) and two heavy chains (blue and yellow) are shown. Glycans between the heavy chains are shown in dark gray.
Author
Katherine Mattaini, Tufts University
- Benetatos, C. A., Y. Mitsuuchi, J. M. Burns, E. M. Neiman, S. M. Condon, G. Yu, M. E. Seipel, G. S. Kapoor, M. G. LaPorte, S. R. Rippin, Y. Deng, M. S. Hendi, P. K. Tirunahari, Y. Lee, T. Haimowitz, M. D. Alexander, M. A. Graham, D. Weng, Y. Shi, M. A. McKinlay, and S. K. Chunduru. 2014. Birinapant (TL32711), a bivalent SMAC mimetic, targets TRAF2-associated cIAPs, abrogates TNF-induced NF-κB activation, and is active in patient-derived xenograft models. Molecular Cancer Therapeutics 13:867-879. ↵


