11.2: Prokaryotic Transcription
- Page ID
- 24826
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)The Lactose Intolerance of Bacteria
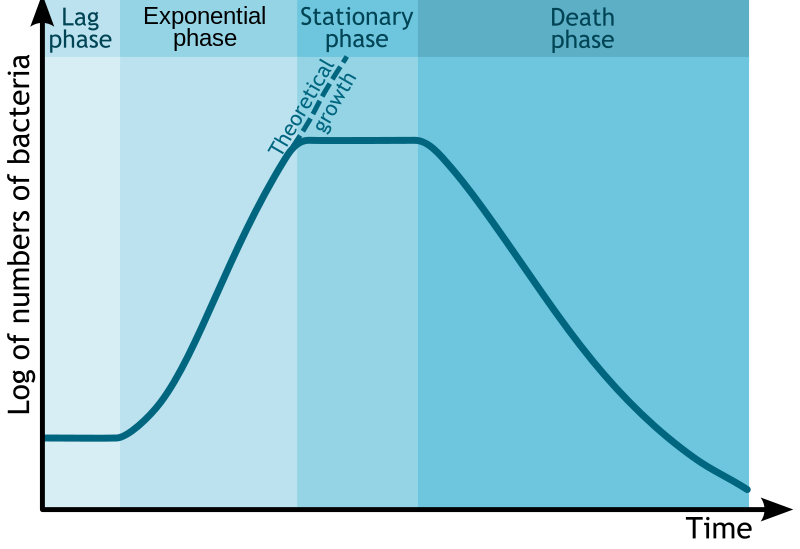
The standard growth kinetics of E. coli are described by the curve. Credit: Michał Komorniczak (CC-BY-SA 3.0)
Glucose is the preferred energy source of cells. François Jacob and Jacques Monod sought to understand how bacteria made decisions to switch between different sugars as sources of energy. Jacob and Monod found that if glucose and lactose were presented as food for bacteria, there would be a biphasic growth pattern.


Credit: CNX OpenStax (CC-BY 4.0)
Jacob and Monod came to understand that the glucose would first be utilized (preferred source) and the lactose would be digested after the depletion of glucose. This occurred because under normal situations the bacteria would not have access to lactose and would waste energy by creating enzymes to digest it. The enzyme β-galactosidase, which is responsible for digesting lactose to the monomers galactose and glucose would only be induced under the conditions of low glucose and high lactose. Monod found that when lactose was the sole sugar, the expression of the enzyme β-galactosidase was induced and displayed a monophasic growth with a delay.
The Lac Operon
Jacob and Monod later found that the genes involved in utilizing lactose were clustered together in proximity under a coordinated control mechanism. This became known as the Lac operon.

A schematic of the Lac Operon. LacZ, LacY, and LacA are transcribed as a single mRNA.
The usage of lactose as a source of energy is preferred by bacteria when glucose is not present. In the presence of abundant glucose, it would be a waste of energy and cellular resources to commit to synthesizing the mRNA and the protein for β-galactosidase. Unless lactose is present, a protein binds to a portion of the Lac promoter referred to as the operator. This repressor protein is encoded by another gene (LacI) outside of the gene cluster. Occasionally, the repressor unbinds to the operator and RNA Polymerase is permitted to transcribe the LacZ gene (β-galactosidase), LacY gene (permease), and LacA gene (acetylase). This “leakiness” of expression is important since the permease protein is needed on the surface of the cell to allow lactose into the cell if it is present in the environment. The presence of lactose causes the repressor to fall off the operator to grant RNA pol access to the DNA. When glucose is low, a protein called CAP (Catabolite Activated Protein) binds to the Lac promoter and works as a recruiter of RNA pol. The coordinated effects of CAP activation and Lac Repressor inactivation yield high transcription of the operon.
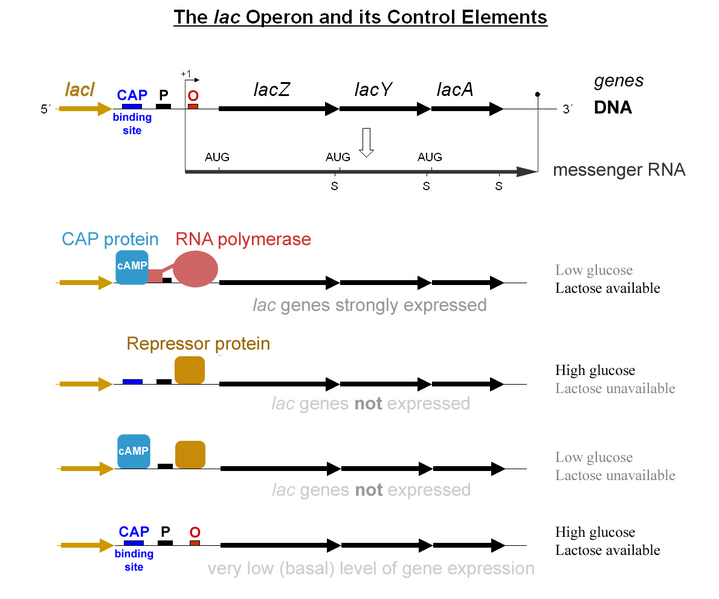
Credit: G3pro (CC-BY 2.0)

LacI bound to 2 DNA operator sequences.
Credit: SocratesJedi (CC-BY-SA 3.0)

Lac Operon Simulation
Launch the simulation below to explore the coordinated activation of the Lac Operon.
LacZ as a Reporter Gene
pUC19 contains LacZ DNA as a reporter gene to illustrate the presence of the functioning gene. Transcription of this gene is driven by the binding site for the RNA Polymerase subunit called σ factor. The σ factor binding site determines the directionality of the RNA polymerase since there is an option of transcribing in 2 directions. The standard σ factor binding site is often denoted as -35 TTGACA…TATAAT -10 from the transcription initiation.
The multiple cloning site within the plasmid provides a convenient location to shuttle a foreign piece of DNA. When no foreign DNA is inserted to this space, the LacZ gene product β-galactosidase is functional. Disruption of the reading frame for this gene likewise disables the functional product from being translated. By using chemical reporters, the integrity of this gene can be confirmed through enzymatic activity.
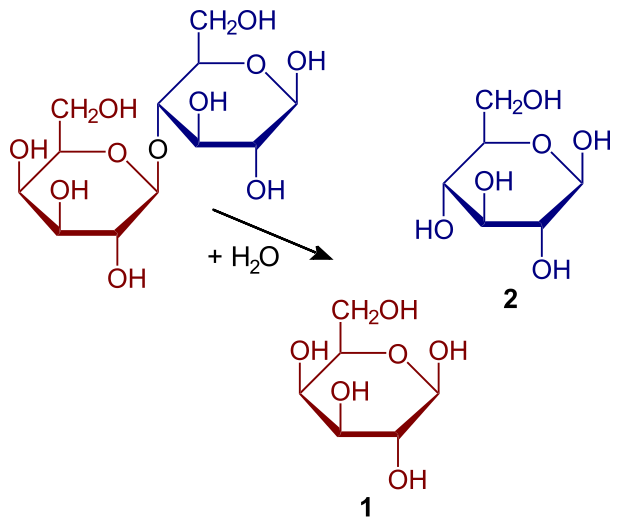
Hydrolysis of lactose to galactose and glucose
Two chemical reporters used to reveal the presence of a functioning LacZ are X-Gal (5-bromo-4-chloro-3-indolyl-β-D-galactoside5-bromo-4-chloro-3-indolyl-β-D-galactoside) and ONPG (orthonitrophenol).
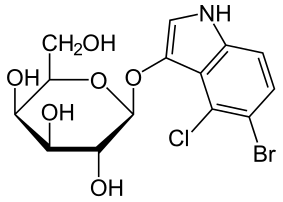
X-Gal (5-bromo-4-chloro-3-indolyl-β-D-galactoside) yields a blue color when cleaved by β-galactosidase

ONPG (orthonitrophenol) yields a yellow color upon cleavage by β-galactosidase
As in the case of the Lac operon, the LacI (repressor protein) will occupy the operator. This operator happens to be overlapping the -35 & -10 sequences. In order to fully activate these genes, the Lac repressor must be removed by binding to a lactose analog. In this case, the chemical IPTG (Isopropyl β-D-1-thiogalactopyranoside) is used since it is a non-cleavable analog that will perpetually bind to the Lac repressor.

IPTG
Blue-White Screening

Blue-White Screening reveals a phenotype of transformed bacteria based on the ability of X-Gal conversion by β-lactamase Credit: Stefan Walkowski (CC-BY-SA 4.0)
References
- JACOB F, MONOD J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961 Jun;3:318-56.


