2.9: Proteins
- Page ID
- 24741
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Proteins are Polymers of Amino Acids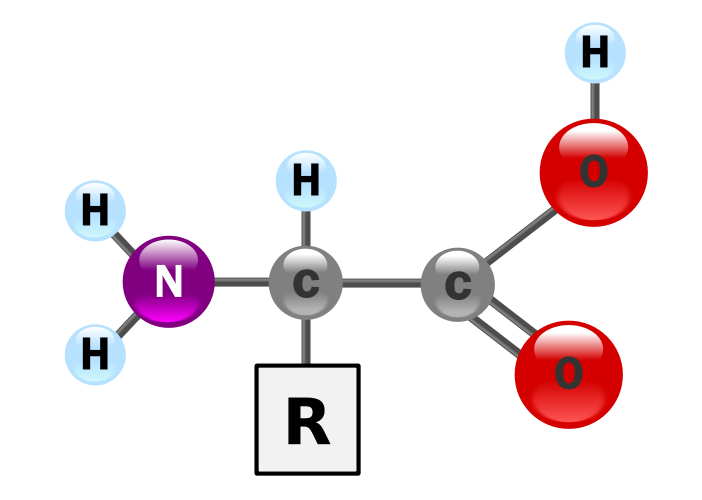
Proteins provide much of the structural and functional capacity of cells. Proteins are composed of monomers called amino acids. Amino Acids are hydrocarbons that have an amino group (-NH2) and an acidic carboxyl group (-COOH). The R group represents a hydrocarbon chain with a modification that alters the properties of the amino acid. 20 universal amino acids are used to construct proteins. The variation in functional groups along the amino acid chain gives rise to the functional diversity of proteins.

20 amino acids and their properties. A 21st amino acid on this table represents the non-universally found selenocysteine.
Monomers bond together through a dehydration synthesis reaction between adjacent amino and carboxyl groups to yield a peptide bond.
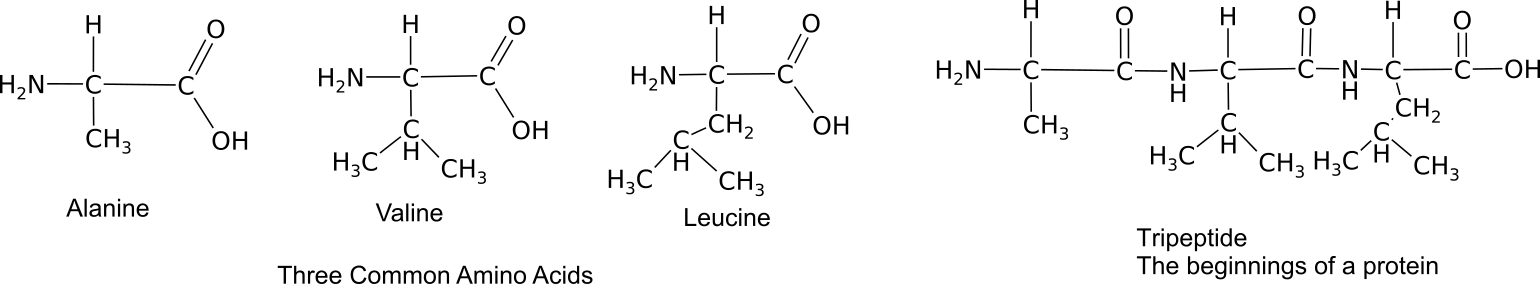
Three amino acids bound into a tripeptide.
How Amino Acids Interact with Each Other and the Environment
Use the following simulation to test how a polypeptide chain with fold based on the type of solution it is in and the composition of the amino acids.
- Protein Folding Simulation (CC BY 4.0 Concord Consortium)
Levels of Structure

- Primary Structure (1°): The sequence of amino acids read from the Amino or N-terminal end of the molecule to the Carboxyl or C-terminal end
- Tyr-Cys-Arg-Phe-Leu-Val-….
- Secondary Structure (2°): local three-dimensional structures that form from interactions of amino acids, like hydrogen bonding
- Alpha Helix – coils occurring from the H-bonds between N-H and C=O groups along the backbone of the protein

Side view of α-helix illustrating H-bonds in magenta between carboxyl oxygen (red) and amine nitrogen (blue)

Top-down view of an α-helix

Side view of ribbon diagram of α-helices traversing a membrane.
- Beta Sheets – laterally connected strands or sheets of amino acids occurring from the H-bonds between N-H and C=O groups along the backbone of the protein
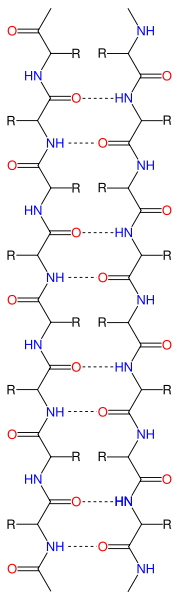

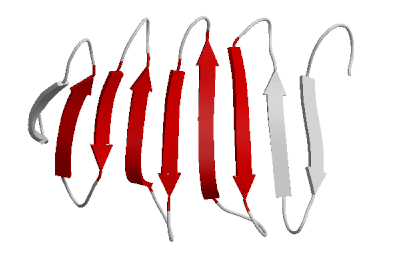
Ribbon diagram of β-sheets
- Tertiary Structure (3°): overall 3-D structure of the peptide chain
- Quaternary Structure(4°): multimeric protein structure from assembling multiple peptide subunits
Diversity of Proteins
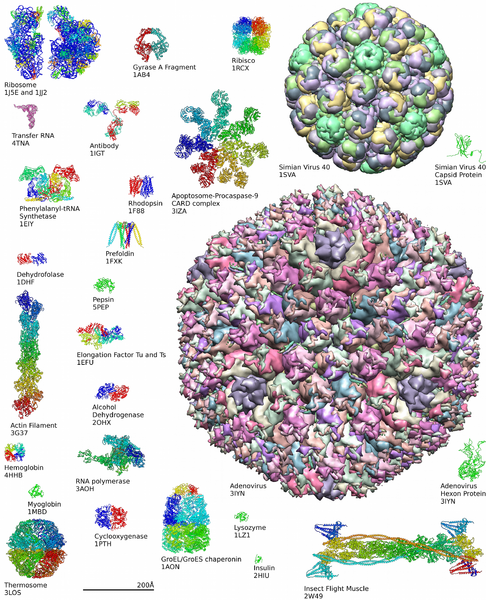
Learn more about the complexity of protein structures at the Protein Data Bank.
Protein Detection (Activity)
Protein Detection Theory:
Proteins can be detected through the use of the Biuret test. Specifically, peptide bonds (C-N bonds) in proteins complex with Cu2+ in Biuret reagent and produce a violet color. A Cu2+ must complex with four to six peptide bonds to produce a color; therefore, free amino acids do not positively react. Long polypeptides (proteins) have many peptide bonds and produce a positive reaction to the reagent. Biuret reagent is an alkaline solution of 1% CuSO4, copper sulfate. The violet color is a positive test for the presence of protein, and the intensity of the color is proportional to the number of peptide bonds in the solution.


Biuret Test
- Examine the table below. Indicate if the sample is a negative control, positive control or an experimental.
- Predict the color change of the solution.
- Formulate a hypothesis about the components of the experimentals.
- Obtain 6 test tubes and number them 1-6.
- Add the materials listed in the table.
- Add 3 drops of Biuret reagent (1.0% CuSO4 with NaOH) to each tube and mix.
- Record the color of the tubes’ contents in Table.
Conclusions about the Urine Samples
Based on the results of the Benedict’s test and the Biuret test, can we make any conclusions?


