Carbohydrates and Glycans 1: Synthesis of a glycan hairpin
- Page ID
- 123582
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)
| Literature-Based Guided Assessment (LGA) | Synthesis of a glycan hairpin |
Instructors: Email hjakubowski@csbsju.edu for answers
Introduction
Glycans can form complex 3D folding shapes as do proteins, but less is known about how they do so. A recent paper demonstrates that glycan strands can fold hairpins reminiscent of β-turns in proteins. This LGA is derived entirely from data, images and figures from this reference:
Fittolani, G., Tyrikos-Ergas, T., Poveda, A. et al. Synthesis of a glycan hairpin. Nat. Chem. 15, 1461–1469 (2023). https://doi.org/10.1038/s41557-023-01255-5. Creative Commons Attribution 4.0 International License. http://creativecommons.org/licenses/by/4.0/.
First, let's review the beta structure in protein including β-turns.
Answer the following questions about Figure A below.

a. Are the beta cartoons in the top part of the panel parallel or antiparallel?
b. The β-turn is shown in blue. Are both turns possible as shown in the diagram with a normal continuous peptide?
c. A synthetic peptide with Gly-CHDA (cis-1,2-cyclohexanedicarboxylic acid) can form a beta-turn with two parallel strands. Draw dashed lines to show hydrogen bonds that stabilize the turns and also the parallel strands in the bottom Lewis structure.
- Answer
-
dkf
Before we consider beta-hairpins in a glycan, let's review the structure of cellulose. Three strands of cellulose are shown in the image below. They form both intrastrand and interstrand hydrogen bonds. Answer the following questions
a. Draw the Symbol Nomenclature for Glycans (SNFG) cartoon structure for strand with 3-monosaccharide repeat. Are the links α or β? What is the repeating monosaccharide?
b. The image below shows three partial strands of cellulose in a single plane. Draw dotted lines showing hydrogen bonds knowing that:
- the ring O has only one intrastrand H bond
- the C3-OH forms both an intrastand and interstrand H bond
- the C2-OH forms only one intrastrand H bond
- the C6-OH forms both an intrastand and interstrand H bond
c. In which direction are the H atoms covalently bonded to the C atoms in the ring pointing? How might this affect other planes of cellulose layers stacking above and below it?
- Answer
-
Given the large number of monomeric monosaccharides, hydroxyl ring substituents, and possible hydrogen bonding interaction patterns, glycans are generally considered very flexible with multiple possible conformations and few empirical or theoretical rules or understandings of how they would fold in complex secondary and tertiary structures. Two such structures are the helical amylose structure arising from the α(1,4) link and the elongated stacked sheets of cellulose ribbons, arising from the β(1,4) linked glucoses. Hence it is hard to image how to synthesize glycan of specific shapes.
In analogy to beta turns in proteins which allow more complex folding patterns, researchers have produced foldable glycan structures that use a similar turn element based on the trisaccharide of the Sialyl LewisX (SLeX) blood group antigen. This tetrasaccharide is found on the ends of glycolipids in white cell membranes where they act as binding motifs as well as surface antigens. Maybe it could be used as a starter for a more complex fold design.
The LewisX trisaccharide has a fucose (SNFG red triangle) and a D-Gal (SNFG yellow circle) connected to D-GlcNAc (SNFG blue square). The structure is shown below (fucose is at the bottom).
a. Draw the SNFG cartoon showing the connections.
b. What are the actual acetal linkages (α, β with numbers) between monomers that are connected covalently?
c. What is the red dotted line between the red ring O and the H?
- Answer
-
Figure \(\PageIndex{4}\) shows an interactive iCn3D model of the Lewis X trisaccharide from MERS-CoV S(6Q05).
Figure \(\PageIndex{18}\): Lewis X trisaccharide from MERS-CoV S (6Q05). (Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i...Lg6NHR5U4ik8UA
Answer the following questions based on the iCn3D model and the static structure.
a. Label the 3 sugars in the trisaccharide.
b. Which atoms in the monomers are connected by the dotted line with the 4.2 Å label?
c. What atom is covalently attached to one of those atoms but is not shown in this and other X-ray structures?
d. What does the dotted line represent and what is its role in the structure?
- Answer
-
Now the research use the LewX trisaccharide and analogs as to which other carbohydrate chains could be added.
Trisaccharide (3mers) I, II and III were made as shown in the figure below Let's just compare 3mer-I and 3mer-II.
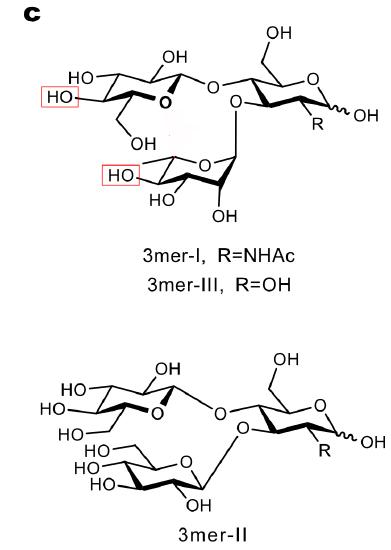
a. What are the differences in 3mer-I and II compared to the LewX trisaccharide?
b. Draw the SNFG representing of the two.
c. Why would the investigators replace the fucose with glucose in 3mer-II?
d. Whey replace the Gal with Glc in both?
- Answer
-
How would test the effects of these changes on the ability of the 3mers to form a β-type turn? Cyrstallography is unlikely, so they used a combination of NMR (which we won't discuss here) and also molecular dynamic (MD) simulations (which we will).
Here are the results of the MD simulations on the 3mers.
Over-imposition of seven representative snapshots (top) and Ramachandran plots for the β-1,4-Glc linkage (bottom) extracted from the MD simulation
a. What do the MD simulations and the Ramachandran plots show? Offer a reason for the results.
b. What is measured in the Ramachandran plots, which are typically discussed when investigating main chain conformations of proteins?
- Answer
-
The next step was to extend the chains at the new terminal Glc residues (that replace the original Gal and Fuc) to create large glycans
A 9mer-I and 9mer-II with the hairpin and two 9er cellulose strands were synthesized and the overall conformations analyzed using MD simulations and NMR. The figure below shows seven representative snapshot from the MD simulations. Explain the results.
- Answer
-
Let's analyze additional results from the MD simulations.
To quantitate the MD simulation results, average distances were measured from pairs of target monomers in the 9mer-I and 9mer-II. The average inter-residue distances are shown in the left panel below. The radius of gyration (Rg) (a measure of the average distance of part of the molecule from the center of mass), is shown on the left. Explain both plots and what the confirm.
- Answer
-
MD suggests that a rigid turn is needed to preserve the closed hairpin conformation. MD suggests that a rigid turn is needed to preserve the closed hairpin conformation
One last analysis combining MD and NMR (a type of measurement called NOE) was done. NOE measures show through-space interactions within a molecule so gives some measure of conformation
A snapshot of the MD simulation of 9mer-I is shown in the figure below with interactions between the two cellulose chains in the 9mer shown as dotted lines. What do the results indicate?
- Answer
-
dfdf


.png?revision=1&size=bestfit&width=396&height=287)