24.3: DNA Recombination
- Page ID
- 15195
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Search Fundamentals of Biochemistry
Homologous Recombination
Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids (usually DNA as in cellular organisms but may be also RNA in viruses). As noted in section 25.2, This process is widely used by cells to accurately repair harmful breaks that occur on both strands of DNA, known as double-strand breaks (DSB), in a process called homologous recombinational repair (HRR). Homologous recombination also produces new combinations of DNA sequences during meiosis, the process by which eukaryotes make gamete cells, like sperm and egg cells in animals. These new combinations of DNA represent genetic variation in offspring, which in turn enables populations to adapt during evolution. Homologous recombination is also used in horizontal gene transfer to exchange genetic material between different strains and species of bacteria and viruses.
Although homologous recombination varies widely among different organisms and cell types, for double-stranded DNA (dsDNA) most forms involve the same basic steps. After a double-strand break occurs, sections of DNA around the 5' ends of the break are cut away in a process called resection. In the strand invasion step that follows, an overhanging 3' end of the broken DNA molecule then "invades" a similar or identical DNA molecule that is not broken. After strand invasion, the further sequence of events may follow either of two main pathways discussed below (see Models); the DSBR (double-strand break repair) pathway or the SDSA (synthesis-dependent strand annealing) pathway. Homologous recombination that occurs during DNA repair tends to result in non-crossover products, in effect restoring the damaged DNA molecule as it existed before the double-strand break.
There are several different ways to repair DSB as illustrated in Figure \(\PageIndex{1}\). The broken (or resected) DNA with a double-stranded break must find and come together (synapse), invade (or intertwine) with the DNA of a donor, that is homologous to it. The repair can then ensue. In somatic cells that undergo mitosis and not meiosis, the preferred donor is the sister chromatid (the copy of one chromosome made during cell division) and not the homologous chromosome (from the diploid cell). Variants include synthesis-dependent strand annealing pathway (SDSA). Other variants include nonhomologous end-joining (NHEJ), microhomology-mediated end-joining (MMEJ), and double Holiday junction (dHJ).
Figure \(\PageIndex{1}\): Model for the repair of DNA double-strand breaks by homologous recombination in somatic cells. Wright et al. J. Biol. Chem. (2018) 293(27) 10524 –10535. Creative Commons Attribution (CC BY 4.0)
When a DNA double-strand break (DSB) occurs in a DNA molecule, a repair can proceed by multiple pathways largely controlled by end resection. NHEJ is capable of repairing unresected or minimally resected DSBs in a template-independent fashion. MMEJ and single-strand annealing (SSA) rely on different extents of homology between the two DSB ends for repair independent of a donor molecule. Homologous recombination proceeds as shown in the figure using a homologous donor DNA. Most of the extended D-loops in somatic cells are disrupted and subsequently repaired by SDSA. The result of the repair by SDSA is always a noncrossover outcome, thus avoiding the loss of heterozygosity produced by somatic crossovers. SDSA occurs by disruption of the extended D-loop and annealing the newly synthesized DNA with the second end of the broken molecule. Alternatively, the newly synthesized strand may invade the second end. The extended D-loop can also undergo second-end capture or invasion to form a double Holliday junction (dHJ). This may either lead to a crossover or a noncrossover outcome. Invasion by the second break end makes dHJ formation and hence crossover outcome more likely for another model for crossover generation. dHJs can be dissolved into noncrossovers by the concerted action of the Sgs1–Top3–Rmi1 complex to migrate the two junctions toward each other and then decatenate the strands of the hemicatenane by the Top3 topoisomerase activity. Each colored line indicates a strand of DNA, and dotted lines represent DNA synthesis.
In the process of homologous recombination, a key intermediate is the Holiday junction, named after Robin Holiday who discovered it. It consists of branched nucleic acid with four double-stranded arms joined. A holiday junction is seen as the crossing of red and blue strands in the middle of Figure 1 labeled Nascent D-loop. Also, one is seen in the Extended D-loop just below it. A double Holiday junction is seen in the middle of the right-hand section. Two views of Holiday junctions are shown in Figure \(\PageIndex{2}\).
|
By Донор - Own work, CC BY-SA 4.0, https://commons.wikimedia.org/w/inde...curid=48470765 |
By Antony-22 - Own work, CC BY-SA 4.0, https://commons.wikimedia.org/w/inde...curid=38557614 |
Figure \(\PageIndex{2}\). Two views of holiday junctions. The left-hand panel shows the primary and secondary sequences and some tertiary (3D) aspects of base-stacking conformational isomers of the Holliday junction. The bases nearest to the junction point determine which stacked isomer dominates.
Figure \(\PageIndex{3}\) shows an interactive iCn3D model of the structure of the Holliday junction intermediate in Cre-loxP site-specific recombination (3CRX).
The alpha carbon backbone of the four Cre recombinase monomers in the tetramer is shown in red. The DNA is nearly planar with a twofold-symmetric DNA intermediate that is similar to a square and stacked Holiday junction for the DNA in the unbound state.
Homologous recombination is conserved across all three domains of life as well as DNA and RNA viruses, suggesting that it is a nearly universal biological mechanism. The discovery of genes for homologous recombination in protists—a diverse group of eukaryotic microorganisms—has been interpreted as evidence that meiosis emerged early in the evolution of eukaryotes. Since their dysfunction has been strongly associated with increased susceptibility to several types of cancer, the proteins that facilitate homologous recombination are topics of active research. Homologous recombination is also used in gene targeting, a technique for introducing genetic changes into target organisms. For their development of this technique, Mario Capecchi, Martin Evans, and Oliver Smithies were awarded the 2007 Nobel Prize for Physiology or Medicine; Capecchi[3] and Smithies[4] independently discovered applications to mouse embryonic stem cells, however, the highly conserved mechanisms underlying the DSB repair model, including uniform homologous integration of transformed DNA (gene therapy), were first shown in plasmid experiments by Orr-Weaver, Szostack, and Rothstein.
Before the beginning of meiosis, the replication of the DNA is required to form sister chromatids, as shown in Step 1 in Figure \(\PageIndex{4}\). Once replicated, the DNA will condense and begin the process of meiosis. As the cells enter metaphase of meiosis, heterologous chromosomes are paired together (Step 2). When the homologous chromosomes are paired, they can under genetic mixing or DNA recombination form new chromosomal arrangements that are unique from the parental chromosomes (Step 3). Once the process of recombination is finished, the heterologous chromosomes are separated into two different daughter cells (Step 4). This is called Meiosis I. At this stage the chromosomes have been reduced from the diploid to the haploid state, however, each chromosome set is still paired with its sister chromatid and needs to undergo a second round of cell division to produce the final set of four gametes (Step 5). This is called Meiosis II and results in the formation of four haploid gametes that are genetically unique.
During the process of meiosis, cell division is used to create the gametes or reproductive cells of an organism (the egg and the sperm cells). Meiosis results in the reduction of the genome from the 2n or diploid state to the 1n or haploid state. As you can see in Figure 25.3.1, the process of meiotic division results in the generation of four genetically unique haploid cells and involves the pairing of heterologous chromosomes during metaphase of meiosis. In humans, the meiotic process results in four viable sperm cells in the male and a single viable egg in the female. The other three cells produced in the female during the meiotic division are termed polar bodies and are very small and do not contain enough cytoplasmic components to survive. They get reabsorbed into the body. In either case, the resulting egg or sperm cell each carries a single copy of the genome and is in the haploid state.
It is during the process of meiosis that homologous recombination occurs in a controlled manner to introduce genetic variation into the resulting gametes. As a result, each egg and sperm cell has a unique genetic makeup that is a mixture of both parental copies of the genome.
Proper segregation during meiosis requires that homologs be connected by the combination of crossovers and sister chromatid cohesion. To generate crossovers, numerous double-strand breaks (DSBs) are introduced throughout the genome by the conserved Spo11 endonuclease. DSB formation and its repair are then highly regulated to ensure that homologous chromosomes contain at least one crossover and that no DSBs remain before meiosis I segregation. The synaptonemal complex (SC) is a meiosis-specific structure formed between homologous chromosomes during prophase that promotes DSB formation and biases the repair of DSBs to homologous chromosomes rather than back to the sister chromatids, ensuring that genetic recombination occurs. Synapsis, the pairing of homologous chromosomes, occurs when a particular recombination pathway is successful in establishing stable interhomolog connections.
Formation of the Synaptonemal Complex
In the 1950s, electron microscopists discovered an evolutionarily conserved, meiosis-specific structure formed between homologous chromosomes, unique to prophase I, called the SC, as shown in Figure \(\PageIndex{5}\). The SC physically connects homologs during prophase I and is removed before metaphase I, when homologs are connected instead by the combination of crossovers and sister chromatid cohesion. What is the function of the SC? Decades of research have shown that this elaborate chromosomal structure is critical for the regulation of recombination, the process by which crossovers are generated.
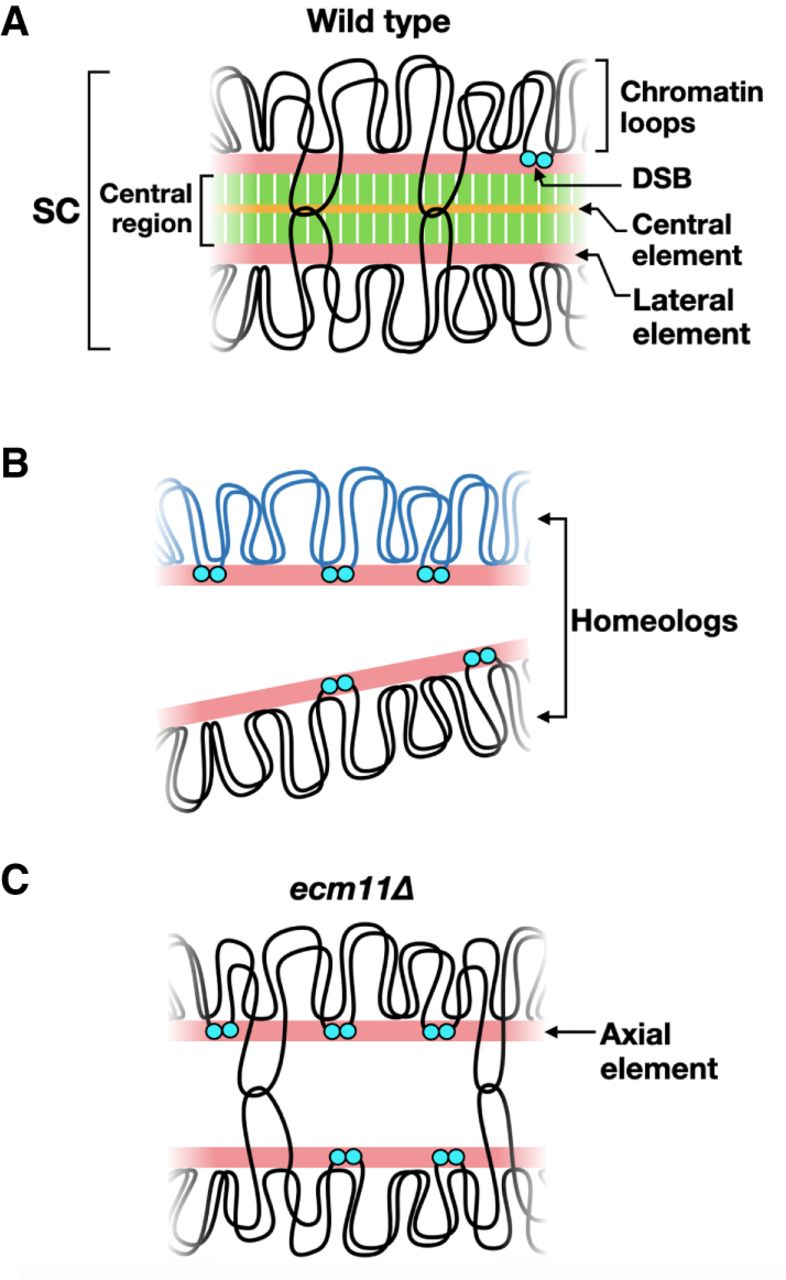
Panel (A) shows the synaptonemal complex. When chromosomes form synapses, recombination intermediates contain double Holliday junctions (shown by intersecting loops). When cells exit the pachytene stage, the stage of meiotic prophase when chromosomes fully synapse, Holliday junctions are resolved to form crossovers, and the SC is disassembled. DSB formation is greatly reduced by synapsis but is not completely abolished until cells exit pachynema.
Panel (B) shows the sequence diversity between homologous chromosomes largely inhibits recombination and synapsis, resulting in persistent DSB formation.
Panel (C) shows that in the absence of the central element, the transverse filament is not assembled, resulting in chromosomes that lack the central region. Recombination intermediates containing double Holliday junctions are still formed, but DSB formation is not down-regulated. Image from:
SC formation begins with the condensation of sister chromatids along meiosis-specific protein cores to make axial elements. Axial elements from homologous chromosomes are “zippered” together by the insertion of the central region. (Note that after synapsis, axial elements are called lateral elements (Panel A above). The central region is comprised of (1) transverse filaments located perpendicular to the lateral elements, and (2) the central element, which runs parallel to the lateral elements midway through the central region.
Assembly of the SC is initiated in the early stage of meiotic prophase I, which is commonly divided into five substages (leptotene, zygotene, pachytene, diplotene, and diakinesis). For proper assembly of the SC followed by the correct pairing of the homologous chromosome, lateral elements (LEs), which are composed of two main proteins (SYCP2 and SYCP3) should be formed along each chromosome at the initial stage, during leptotene. Later the two LEs associate with the linker part, known as the transverse filaments (TFs). TFs are primarily composed of the protein SYCP1. The central element (CE), which is composed of SYCE1, SYCE2, SYCE3, and TEX12, then connects to the LEs through the TFs, as shown in Figure \(\PageIndex{6}\).
The lateral elements complete their pairing during the zygotene stage leading to the formation of the tripartite SC structure seen during the pachytene stage of the first meiotic prophase as shown in Figure \(\PageIndex{7}\) and Figure \(\PageIndex{8}\). This occurs in both males and females during gametogenesis.
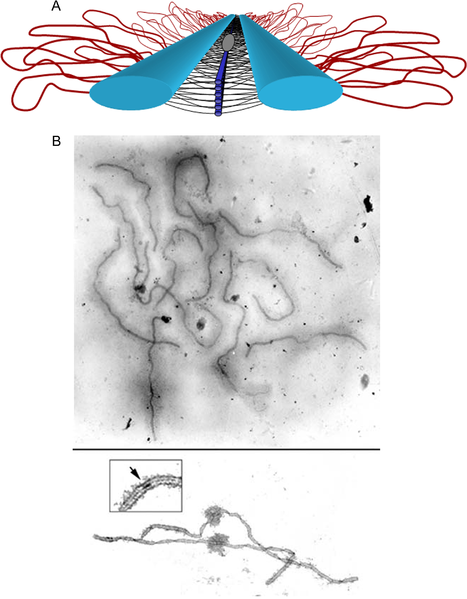
Panel (A) shows a model of the SC. Lateral elements (light blue rods) of homologous chromosomes align and synapse together via a meshwork of transverse filaments (black lines) and longitudinal filaments (dark blue rods). The longitudinal filaments are collectively referred to as the “central element” of the SC. Ellipsoidal structures called recombination nodules (gray ellipsoid) are constructed on the central region of the SC. As their name implies, recombination nodules are believed to be involved in facilitating meiotic recombination (crossing over). The chromatin (red loops) of each homolog is attached to its corresponding lateral element. Because there are two “sister chromatids” in each homolog, two loops are shown extending laterally from each point along a lateral element.
Panel (B) Top shows a set of tomato SCs. Chromatin “sheaths” are visible around each SC showing two tomato SCs. The chromatin has been stripped from the SCs, allowing the details of the SC to be observed. Each SC has a kinetochore (“ball-like” structure) at its centromere. Recombination nodules, ellipsoidal structures found on the central regions of SCs, mark the sites of crossover events (see inset).
Zygotene is the sub-stage where synapsis between homologous chromosomes begins. It is also known as zygonema. This synapse can form up and down the chromosomes allowing numerous points of contact called 'synaptonemal complex', this can be compared to a zipper structure, due to the coils of chromatin. The SC facilitates synapsis by holding the aligned chromosomes together. After the homologous pairs synapse they are either called tetrads or bivalents. Bivalent is more commonly used at an advanced level as it is a better choice due to similar names for similar states (a single homolog is a 'univalent', and three homologs are a 'trivalent').
Once the synapse is formed it is called a bivalent (where a chromatid of one pair is synapsed/attached to the chromatid in a homologous chromosome and crossing over can occur. Subsequently, the synapses snap completing the crossing over of the genetic information. As a result, the variation in genetic material has increased significantly, because up and down the chromosome there has been an exchange of the mother and father's genetic material. The two sister chromatids separate from each other, but the homologous chromosomes remain attached. This makes the complex look much thicker. The SC is complete, allowing chiasma to form. This is what allows the crossing over alleles to occur as this is a process that only happens over a small region of the chromosomes.
The chiasma is a structure that forms between a pair of homologous chromosomes by crossover recombination and physically links the homologous chromosomes during meiosis as shown in Figure \(\PageIndex{9}\). Chiasmata are essential for the attachment of the homologous chromosomes to opposite spindle poles (bipolar attachment) and their subsequent segregation to the opposite poles during meiosis I.
Mechanism of Homologous Recombination
Meiotic recombination is a tightly regulated process that is triggered by the programmed induction of DNA double-strand breaks (DSBs). Once formed, the ends of the DSBs are nucleolytically processed to generate 3′ single-stranded DNA (ssDNA) tails. Meiotic recombination factors then engage these ssDNA tails to form a nucleoprotein ensemble capable of locating DNA homology in the chromosome homolog and mediating invasion of the homolog to form a DNA joint called a displacement loop or D-loop. The 3′ end of the invading strand is extended by DNA synthesis, followed by the pairing of the non-invading 3′ single-stranded tail with the displaced ssDNA strand in the enlarged D-loop (second-end capture). After DNA synthesis and DNA ligation, a double Holliday Junction (dHJ) intermediate is formed. Resolution of the dHJ intermediate can result in crossover recombinants that harbor a reciprocal exchange of the arms of the homologous chromosomes.
Genetic studies have revealed that meiotic DSBs arise via the action of a protein ensemble that harbors the Spo11 protein, which bears homologous to archaeal Topo VIA, the catalytic subunit of a type II topoisomerase. Indeed, studies in S. cerevisiae, S. pombe, and M. musculus have shown that Spo11 becomes covalently conjugated to the 5′ ends of DNA through a tyrosine residue proposed to be the catalytic center of topoisomerase function. Thus, mutations in the putative catalytic tyrosine residue of Spo11 engender the same phenotype as spo11 deletion in S. cerevisiae, S. pombe, A. thaliana, and M. musculus. All these observations suggest that Spo11 is directly involved in catalyzing DSB formation to trigger meiotic recombination. Figure \(\PageIndex{10}\) provides an overview of this process.
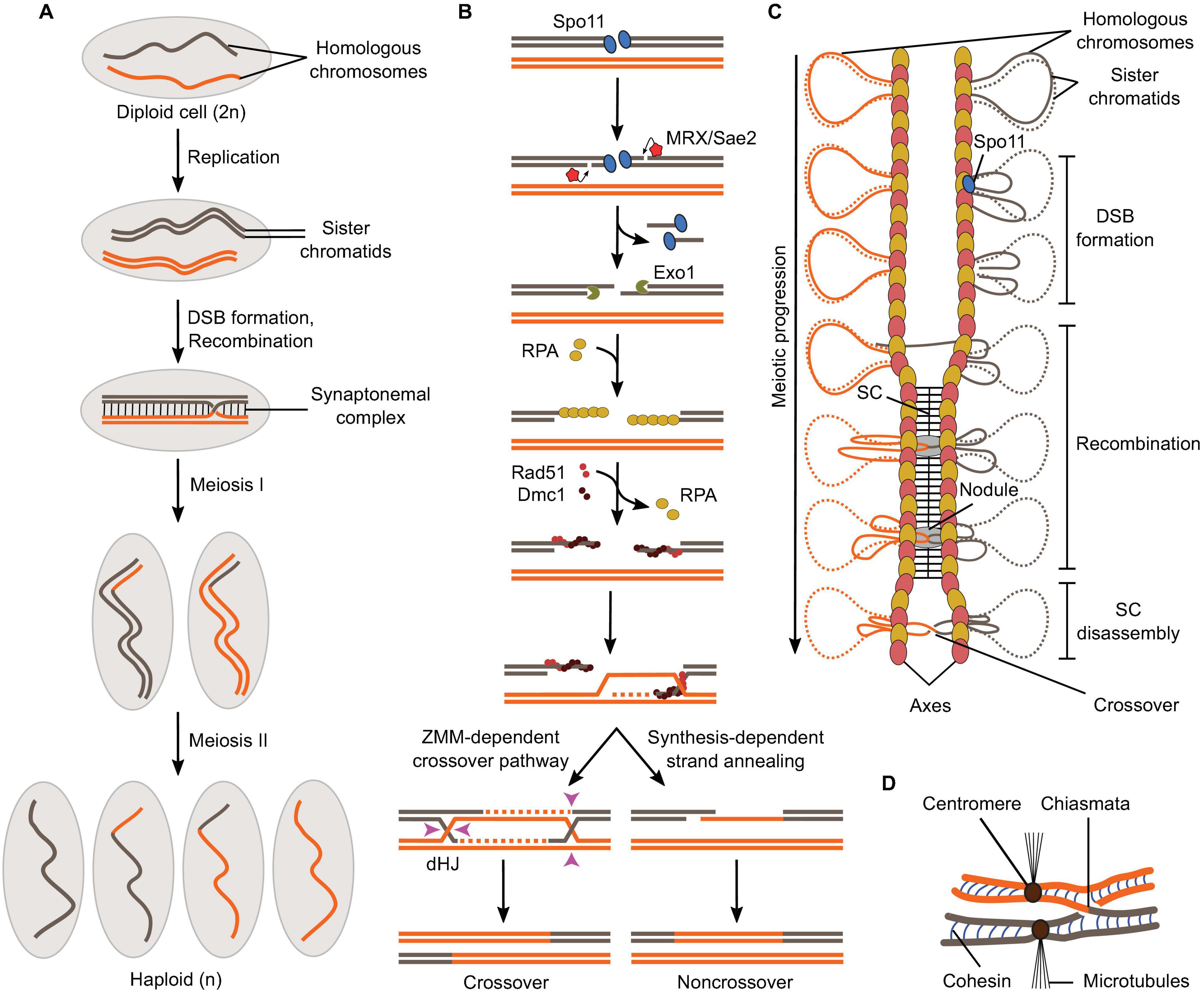
Panel (A) shows a schematic of the formation of haploid gametes from a diploid cell with a single pair of homologous chromosomes. DSB formation and recombination promote homolog pairing and lead to the exchange of chromosomal fragments (crossovers) in the context of synapsed chromosomes.
Panel (B) shows meiotic recombination is initiated by Spo11-mediated DSB formation and leads to the formation of crossovers via a ZMM-dependent double Holliday Junction (dHJ) resolution pathway or non-crossovers by synthesis-dependent strand annealing.
Panel (C) shows the relationships between meiotic recombination and higher-order chromosome structure. DSB formation happens in the context of the loop-axis structure. As recombination progresses, the SC polymerizes between the axes and is disassembled before chromosome segregation. Axis proteins Red1 (red ovals) and Hop1 (yellow ovals) are shown.
Panel (D)shows that in metaphase I, homologs are held together through chiasmata and sister chromatid cohesion. Image from:
Following break formation, Spo11 remains covalently attached to the 5′-strands at both DNA ends and is released by an endonucleolytic cleavage reaction mediated by MRX (Mre11, Rad50, and Xrs2) and Sae2, which liberates Spo11 attached to a short oligonucleotide (Fig. 25.3.7B). The 5′-strands are further resected by 5′-3′ exonucleases to produce long single-stranded tails, which are coated with the ssDNA-binding protein, RPA. RPA is then replaced by recombinases Rad51 and Dmc1 that form a nucleoprotein filament and search for sequence similarity preferentially located on the homologous chromosome, producing D-loop structures. Following DNA synthesis using the homolog as a repair template, the recombination structures experience one of two main outcomes (Fig. 25.3.7B). The invading strand can be ejected from the donor by the action of helicases, which provides an opportunity for the DNA ends to re-anneal. This process is referred to as synthesis-dependent strand annealing (SDSA) and produces non-crossovers, that is, products not associated with reciprocal exchanges of chromosome fragments, but with the local transfer of genetic information from the repair template to the broken molecule (gene conversion). Alternatively, recombination structures are stabilized by the “ZMM” family of proteins and channeled through a pathway that produces mostly crossovers. Here, both ends of the break engage the donor to form a double Holliday Junction intermediate, which is resolved through a crossover-specific pathway that involves MutLγ and Exo1.
Every aspect of meiotic recombination is tied to the structural organization of the chromosomes (Fig. 25.3.7C). Early in the meiotic prophase, chromosomes organize as a series of DNA loops that are anchored along a nucleoprotein axis. DSB formation happens in the context of this loop-axis structure. As recombination progresses, polymerization of a proteinaceous structure called the synaptonemal complex (SC) initiates between the two axes and elongates along their entire length. Recombination proceeds within the SC, inside a nodule embedded between the axes. After recombination is completed, the SC disassembles and crossovers, now cytologically visible as chiasmata, provide physical connections between the homologs until their segregation at anaphase (Fig. 25.3.7D).
References
1. Hollingsworth, N.M (2020) A new role for the synaptonemal complex in the regulation of meiotic recombination. Genes and Dev. 34: 1562-1564. Available at: http://genesdev.cshlp.org/content/34/23-24/1562.full
2. Seo, E.K., Choi, J.Y., Jeong, J-H., Kim Y-G, Park, H.H. (2016) Crystal structure of C-terminal coiled-coil domain of XYCP1 reveals the non-canonical anti-parallel dimeric structure of transverse filament at the synaptonemal complex. PLOS one: DOI 10.1371. Available at: https://www.researchgate.net/publication/306394048_Crystal_Structure_of_C-Terminal_Coiled-Coil_Domain_of_SYCP1_Reveals_Non-Canonical_Anti-Parallel_Dimeric_Structure_of_Transverse_Filament_at_the_Synaptonemal_Complex
3. Wikipedia contributors. (2021, April 18). Synaptonemal complex. In Wikipedia, The Free Encyclopedia. Retrieved 02:07, August 7, 2021, from https://en.Wikipedia.org/w/index.php?title=Synaptonemal_complex&oldid=1018542057
4. Wikipedia contributors. (2021, July 21). Homologous recombination. In Wikipedia, The Free Encyclopedia. Retrieved 02:17, August 7, 2021, from https://en.Wikipedia.org/w/index.php?title=Homologous_recombination&oldid=1034703880
5. School of Biomedical Wiki (Accessed Aug 2021) Meiosis Prophase I. Available at: https://teaching.ncl.ac.uk/bms/wiki/index.php/Meiosis_prophase_1
6. Hirose, Y., Suzuki, R., Ohba, T., Hinohara, Y., Matsuhara, H., Yoshida, M., Itabashi, Y., Murakami, H., and Yamamoto, A. (2011) Chaismata Promote Monopolar Attachment of Sister Chromatids and Their Co-Segregation Toward the Proper Pole during Meiosis I. PLoS Genet. 7(3):e1001329. Available at: https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1001329
7. Yeh, H-Y., Lin, S-W., Wu, Y-C., Chan, N-L., and Chi P. (2017) Functional Characterization of the Meiosis-Specific DNA Double-Strand Break Inducing Factor, SPO-11 from C. elegans. Scientific Reports 7:2370. Available at: https://www.nature.com/articles/s41598-017-02641-z#rightslink
8. Yadav, V.K., and Bouuaert, C.C. (2021) Mechanism and Control of Meiotic DNA Double-Strand Break Formation in S. cerevisiae. Front. Cell Dev. Biol. 9:642737. Available at: https://www.frontiersin.org/articles/10.3389/fcell.2021.642737/full




.png?revision=1&size=bestfit&width=356&height=314)