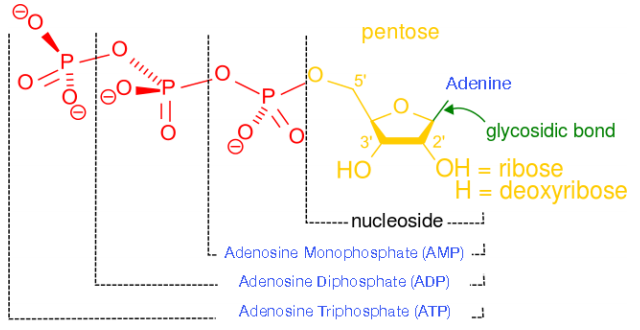
2.6: Cellular Phosphorylations
- Page ID
- 1709
Formation of triphosphates is essential to meet the cell’s immediate energy needs for synthesis, motion, and signaling. In a given day, an average human being uses more than their body weight in triphosphates. Since triphosphates are the “currency” that meet immediate needs of the cell, it is important to understand how triphosphates are made. There are three phosphorylation mechanisms – 1) substrate level; 2) oxidative; and 3) photophosphorylation. We consider them here individually.
Substrate Level Phosphorylation
The easiest type of phosphorylation to understand is that which occurs at the substrate level. This type of phosphorylation involves the direct synthesis of ATP from ADP and a reactive intermediate, typically a high energy phosphate-containing molecule. Substrate level phosphorylation is a relatively minor contributor to the total synthesis of triphosphates by cells. An example substrate phosphorylation comes from glycolysis.
\[\ce{Phosphoenolpyruvate (PEP) + ADP <=> Pyuvate + ATP}\]
This reaction has a very negative \(ΔG^{o'}\) (-31.4 kJ/mol), indicating that the PEP contains more energy than ATP, thus energetically favoring ATP’s synthesis. Other triphosphates can be made by substrate level phosphorylation, as well. For example, GTP can be synthesized by the following citric acid cycle reaction
\[\ce{Succinyl-CoA + GDP + P_i <=> Succinate + GTP + CoA-SH}\]
Triphosphates can be interchanged readily in substrate level phosphorylations catalyzed by the enzyme Nucleoside Diphosphate Kinase (NDPK). A generalized form of the reactions catalyzed by this enzyme is as follows:
\[\ce{XTP + YDP <=> XDP + YTP}\]
where \(\ce{X}\) can be adenosine, cytidine, uridine, thymidine, or guanosine and \(\ce{Y}\) can be any of these as well. Last, an unusual way of synthesizing ATP by substrate level phosphorylation is that catalyzed by adenylate kinase
\[\ce{2 ADP <=> ATP + AMP}\]
This reaction is an important means of generating ATP when the cell doesn’t have other sources of energy. The accumulation of AMP resulting from this reaction activates enzymes, such as phosphofructokinase, of glycolysis, that will catalyze reactions to give the cell additional, needed energy.
Electron Transport/Oxidative Phosphorylation
Mitocohondria are called the power plants of the cell because most of a cell’s ATP is produced there, in a process referred to as oxidative phosphorylation. The mechanism by which ATP is made in oxidative phosphorylation is one of the most interesting processes in all of biology. It has three primary considerations. The first is electrical – electrons from reduced energy carriers, such as NADH and FADH2, enter an electron transport system via protein complexes containing iron. As seen in the figure on the following page,electrons move from one complex to the next, not unlike the way they might move through an electrical circuit.
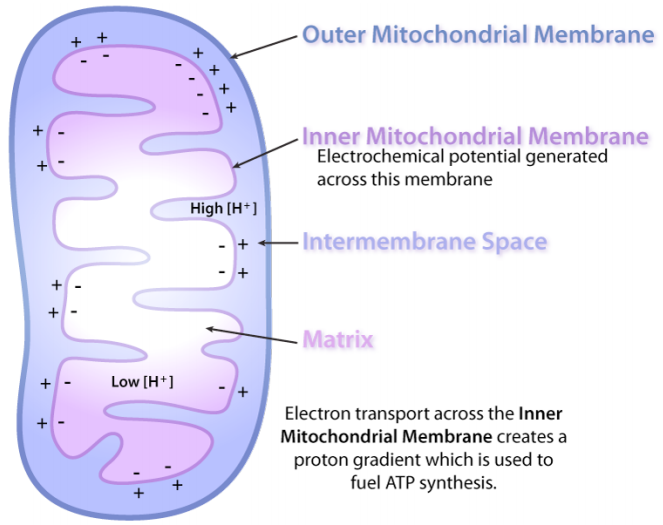
The next consideration arises as a secondary phenomenon. When electrons pass through complexes I, III, and IV, protons are moved from the mitochondrial matrix (inside of mitochondrion) and deposited in the intermembrane space (between the inner and outer membranes of the mitochondrion). The effect of this redistribution is to increase the electrical and chemical potential across the membrane. Students may think of the process as “charging the battery.”
Just like a charged battery, the potential arising from the proton differential across the membrane can be used to do things. This is the third consideration. In the mitochondrion, the “thing” that the proton gradient does is create ATP from ADP and Pi (inorganic phosphate). This process requires energy and is accomplished by movement of protons through a protein complex in the inner mitochondrial membrane. The protein complex is an enzyme that has several names, including Complex V, PTAS (Proton Translocating ATP Synthase), and ATP Synthase. Central to its function is the movement of protons through it (from outside back into the matrix). Protons will only move through ATP Synthase if their concentration is greater outside the inner membrane than in the matrix.
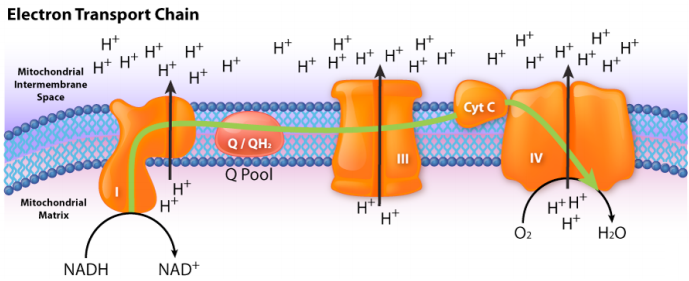
In summary, the electron transport system charges the battery for oxidative phosphorylation by pumping protons out of the mitochondrion. The intact inner membrane of the mitochondrion keeps the protons out, except for those that re-enter through ATP Synthase. The ATP Synthase allows protons to re-enter the mitochondrial matrix and harvests their energy to make ATP.
ATP Synthase
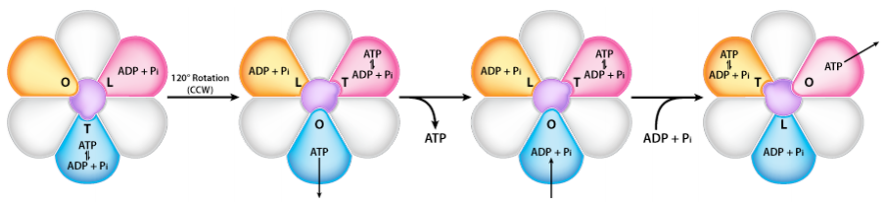
The ATP Synthase itself is an amazing nanomachine that makes ATP using a gradient of protons flowing through it from the intermembrane space back into the matrix. It is not easy to depict in a single image what the synthase does. The figure at the right illustrates the multi-subunit nature of this membrane protein, which acts like a turbine at a hydroelectric dam. The movement of protons through the ATP Synthase causes it to spin like a turbine, and the spinning is necessary for making ATP.
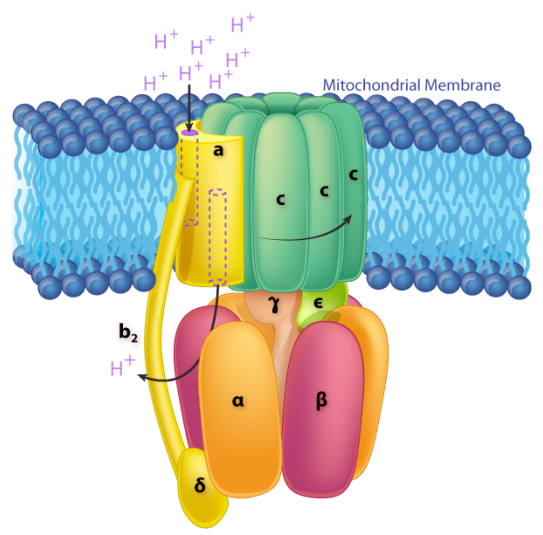
In ATP Synthase, the spinning component is the membrane portion (c ring) of the F0 stalk. The c ring proteins are linked to the gamma-epsilon stalk, which projects into the F1 head of the mushroom structure. The F1 head contains the catalytic ability to make ATP. The F1 head is hexameric in structure with paired alpha and beta proteins arranged in a trimer of dimers. Movement of the gamma protein inside the alpha-beta trimer causes each set of beta proteins to change structure slightly into three different forms called Loose, Tight, and Open (L,T,O). Each of these forms has a function. The Loose form binds \(\ce{ADP}\) and \(\ce{P_i}\). The tight form “squeezes” them together to form the ATP. The open form releases the ATP into the mitochondrial matrix. Thus,as a result of the proton excess in the intermembrane space, ATP is made.
Photophosphorylation
The third type of phosphorylation to make ATP is found only in cells that carry out photosynthesis. This process is similar to oxidative phosphorylation in several ways. A primary difference is the ultimate source of the energy for ATP synthesis. In oxidative phosphorylation, the energy comes from electrons produced by oxidation of biological molecules. In the case of photosynthesis, the energy comes from the light of the sun.
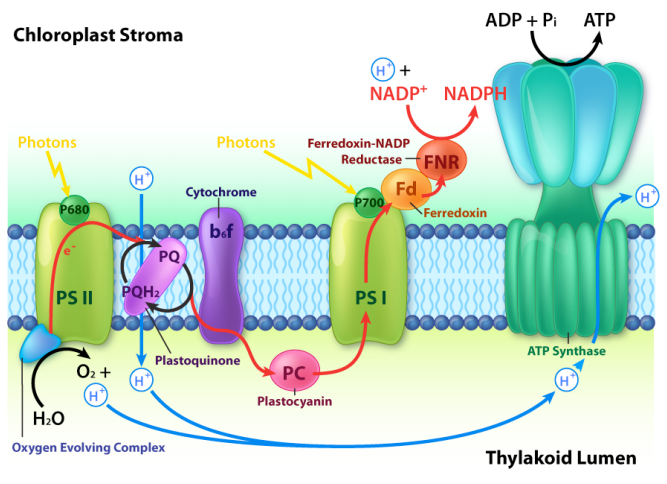
Photons from the sun interact with chlorophyll molecules in reaction centers in the chloroplasts of plants or membranes of photosynthetic bacteria. A schematic of the process is shown above. The similarities of photophosphorylation to oxidative phosphorylation include:
- an electron transport chain
- creation of a proton gradient
- harvesting energy of the proton gradient by making ATP
with the help of an ATP synthase. Some of the differences include:
- the source of the electrons – \(\ce{H2O}\) for photosynthesis versus \(\ce{NADH/FADH2}\) for oxidative phosphorylation
- direction of proton pumping – into the thylakoid space of the chloroplasts versus outside the matrix of the mitochondrion
- movement of protons during ATP synthesis – out of the thylakoid space in photosynthesis versus into the mitochondrial matrix
- nature of the terminal electron acceptor – \(\ce{NADP^{+}}\) in photosynthesis versus \(\ce{O2}\) in oxidative phosphorylation.
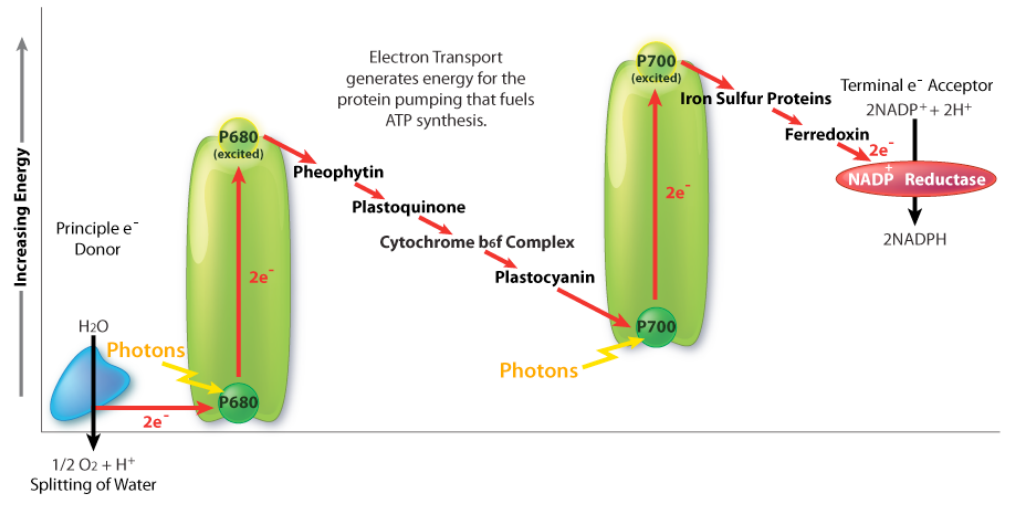
Electron Transport in Chloroplasts vs. Mitochondria
In some ways, the movement of electrons in chloroplasts during photosynthesis is opposite that of electron transport in mitochondria. In photosynthesis, water is the source of electrons and their final destination is \(\ce{NADPH}\). In mitochondria, \(\ce{NADH/ FADH2}\) are electron sources and \(\ce{H2O}\) is their final destination.
How do biological systems get electrons to go both ways? It would seem to be the equivalent of going to and from a particular place while always going downhill, since electrons will move according to potential. The answer is the captured energy of the photons, which elevates electrons in photosynthesis to an energy where they move “downhill” to their \(\ce{NADPH}\) destination in a Z-shaped scheme. The movement of electrons through this scheme in plants requires energy from photons in two places to “lift” the energy of the electrons sufficiently. Last, it should be noted that photosynthesis actually has two phases, referred to as the light cycle (described above) and the dark cycle, which is a set of chemical reactions that captures \(\ce{CO2}\) from the atmosphere and “fixes” it, ultimately into glucose. The dark cycle is also referred to as the Calvin Cycle.


