18.4: Microtubules are Dynamic Structures Composed of Tubulin Monomers
- Page ID
- 89022
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Microtubules assemble from dimers of \(\alpha\)−tubulin and \(\beta\)−tubulin monomers. After their formation, \(\alpha / \beta\)−tubulin dimers add to a growing, or plus end (+end), fueled by GTP hydrolysis (see Figure 18.2). Disassembly at the −end of microtubules powers the changes in cell shape or in the separation and movement of chromatids to opposite poles of cells during mitosis or meiosis.
Isolated single microtubules were shown to grow by addition to one end and to disassemble at the opposite end, thus distinguishing the +ends and −ends. Find a summary of evidence for microtubule polarity in the following link.
323-2 Demonstration of the Polarity & Dynamics of Microtubules
Microtubules in most cells (other than when they are dividing) can seem disordered. In nondividing (interphase) animal cells, they tend to radiate from centrioles without forming discrete structures. But as cell division approaches, microtubules reorganize to form spindle fibers. The reorganization is nucleated from centrioles in animal cells and from a more amorphous MicroTubule Organizing Center (MTOC) in plant cells. A typical centriole (or basal body) has a “nine-triplet” microtubule array, as seen in electron-micrograph cross sections (Figure 18.4).
18.4.1 The Two Kinds of Microtubules in Spindle Fibers
The spindle fibers of mitosis and meiosis are made up of kinetochore microtubules and polar microtubules. The former use chemical energy to pull duplicated chromatids apart; the latter use chemical energy to separate daughter cells during cytokinesis.
18.4.1.a Kinetochore Microtubules
Duplicated chromosomes condense in prophase of mitosis and meiosis, forming visible paired chromatids attached at their centromeres. Specific proteins associate with centromeres to make a kinetochore during condensation. As the spindle apparatus forms, some spindle fibers attach to the kinetochore; these are the kinetochore microtubules. By metaphase, bundles of kinetochore microtubules stretch from the kinetochores at the cell center to the centrioles at opposite poles of a dividing animal cell, as drawn in Figure 18.5.

The +ends of kinetochore microtubules are in fact at the kinetochores, where these fibers assemble! During anaphase, kinetochore microtubules disassemble and shorten at their −ends (at the centrioles in animals or at the MTOCs in plant cells). The forces generated by these activities pull against the centromeres of the chromatids, separating them and then, drawing what are now daughter chromosomes to the opposite poles of the dividing cell. The role of kinetochore microtubule disassembly at the centrioles (i.e., at their −ends) was shown in a clever experiment in which a tiny laser beam was aimed into a cell at the spindle fibers attached to the kinetochore of a pair of chromatids. (See Figure 18.6 below as well as an animation of these events at the link that follows.)

18.4.1.b Polar Microtubules
Spindle-fiber polar microtubules extend from centrioles/MTOCs at opposite poles, toward the center of dividing cells. But instead of binding to kinetochores, they overlap at the center of the dividing cells. While the kinetochore microtubules are tugging apart the paired chromatids at the center of the cell, the polar microtubules are sliding past one another in opposite directions, pushing apart the poles of the cell. In this case, dynein motor proteins attached to microtubules (illustrated in Figs. 18.5 and 18.6) hydrolyze ATP to power microtubule sliding. The dynein motors on the microtubules at one pole of the cell in effect “walk” along overlapping microtubules extending from the opposite pole.
18.4.2 Microtubules in Cilia and Flagella
The microtubules of cilia or flagella emerge from a basal body, shown in the electron micrograph at the left in Figure 18.7.

Basal bodies, organized as nine-triplet microtubule rings, are structurally similar to centrioles. It turns out that eukaryotic flagella and cilia can be stripped from their cells in a very highspeed blender (not the kind you may have in your kitchen!). Treatment with detergents dissolves their membranes, leaving behind axonemes with a 9 + 2 array of microtubules. While the formation of cilia and flagella begins at the basal bodies, both structures soon show a typical 9 + 2 arrangement (i.e., nine outer doublets plus two central singlet microtubules) seen in the cross section at the right in the micrograph). Axonemes from cilia and flagella are virtually identical.
Detergents, including those found in shampoo and laundry “detergents”, contain emulsifying chemicals like sodium lauryl sulfate. Why would a detergent be effective in isolating axonemes?
Microtubules and axonemes are shown in greater detail in Figure 18.8 (below). In the inset at the upper left, it is possible to see the tubulin subunits that make up a microtubule polymer in cross section. Each tubule is made up of a ring of thirteen tubulin subunits. In the high magnification, high resolution cross section micrograph of an axoneme at the right in Figure 18.8, the microtubules in the doublets share tubulins but are also composed of thirteen tubulins. When fully formed, the 25 nm diameter microtubules appear to be hollow cylinders. When isolated, they typically come along with the dynein motor proteins and other Microtubule-Associated Proteins (MAPs), indicated in the micrograph on the right (Figure 18.8). These proteins hold microtubules together in an axoneme and play a role in motility.
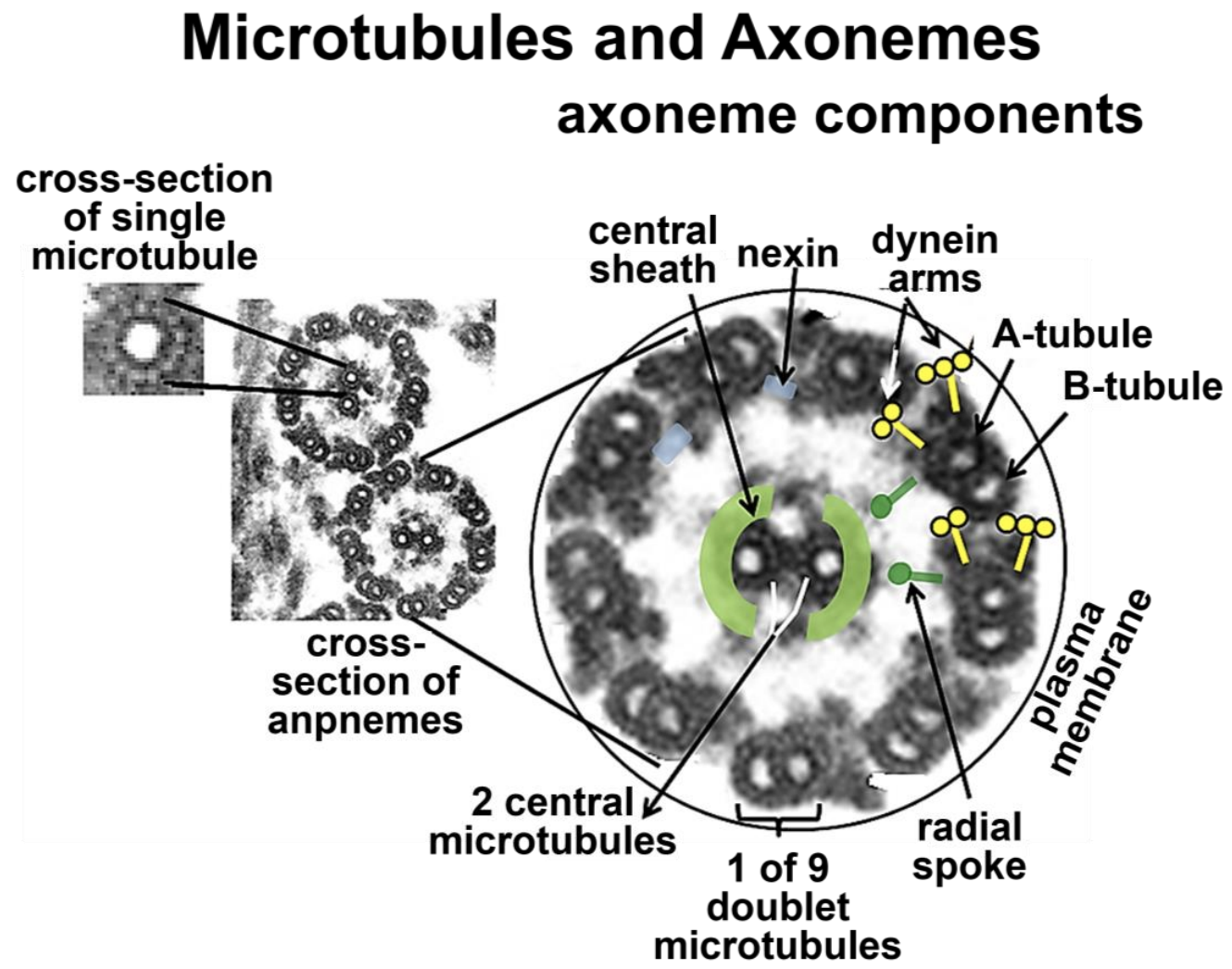
When fully formed, the 25 nm diameter microtubules appear to be hollow cylinders. When isolated, they typically come along with the dynein motor proteins and other MAPs labeled in the micrograph at the right. These proteins hold microtubules together in an axoneme and play a role in motility.
18.4.3 Microtubule Motor Proteins Move Cargo from Place to Place in Cells
Motor proteins, such as dynein and kinesin, are ATPases; they use the free energy of ATP hydrolysis to power intracellular motility. Let’s take a closer look at how these two major motor proteins carry cargo from place to place inside of cells. Organelles are a typical cargo. Examples include vesicles formed at the trans-Golgi face, which contain secretory proteins, pigments, or neurotransmitters. Secretory vesicles move along microtubule tracks to the plasma membrane for exocytosis. Vesicles containing neurotransmitters move from the cell body of neurons, along microtubule tracks in the axons, and reach the nerve ending where they become synaptic vesicles. In a chameleon, pigment vesicles in skin cells disperse or aggregate along microtubule tracks to change skin color to match the background.
Vesicle transport in neurons is well understood. Neurotransmitter vesicles arise from the endomembrane system in neuron cell bodies. ATP-dependent kinesin motor proteins power anterograde movement of the vesicles (from the cell body to the nerve endings). In contrast, ATP-dependent dynein motors (part of a dynactin complex) power retrograde movement of empty vesicles back to the cell body. Motor protein structure and action are shown in Figure 18.9.

325-2 Microtubule Motor Proteins
A fanciful (and not too inaccurate!) animation of a motor protein in action on an axonal microtubule is at this link: Kinesin 'Walks' An Organelle Along a Microtubule. Next let’s look at some elegant studies of isolated axonemes and see what they tell us about microtubule based cell motility.
18.4.4 Demonstrating Sliding Microtubules
Experiments on axonemes that have been isolated from demembranated cilia or flagella confirm the sliding-microtubule mechanism of ciliary and flagellar motility. For example, adding ATP to detached cilia or flagella makes them beat in much the same way as they do when attached to their cells. The phenomenon is easily seen in a light microscope. But isolated axonemes (with their original 9 + 2 microtubule arrangement) also “beat” (after a fashion!) in the presence of ATP! The experiment is illustrated in Figure 18.10 (below).

326-2 9+2 The Microtubule Array in Axonemes that Beat
Selective addition of different detergents removes radial spokes, nexin, and other proteins from the axoneme, causing the microtubules to separate. Dissociated microtubule doublets and central “singlets” can then be observed in the electron microscope. When such separated microtubules are dialyzed to remove the detergents, doublet microtubules reassociate to form sheets, as shown in Figure 18.11.

ATP added to reconstituted microtubule doublets causes the microtubules to separate as the ATP is hydrolyzed. When such preparations are fixed for electron microscopy immediately after adding the ATP, the microtubules are caught in the act of sliding. See this animated in the following link (#327-2).
372-2 Proof of Sliding Microtubules in Flagella and Cilia
328 Bacterial Flagella are Powered by a Proton Gradient
329 The Effects of Different Drugs on Microtubules... and Cancer
18.4.5 The Motor Protein Dynein Enables Axonemes to Bend
Take another look at the cross section of axonemes in Figure 18.9. In the 9 + 2 axoneme of cilia and flagella, dynein arms attached to the A tubules of the outer doublets walk along the B tubules of the adjacent doublet. If the doublets on one side of an axoneme take a walk while those on the other side hold still, the microtubules will slide past one another, and the axoneme (and therefore a cilium or flagellum) will bend. This microtubule sliding is constrained by flexible nexin and radial spoke attachments. Figure 18.12 compares the movements of cilia and flagella.

The differences in flagellar motion (wave-like propeller) and ciliary motion (back-and-forth beat in a single plane) result in part from which microtubules are sliding at a given moment and the nature of their restraint by axoneme proteins.


