17.7: Directing the Traffic of Proteins in Cells
- Page ID
- 89013
All polypeptide proteins have been translated by ribosomes from a sequence of bases in an mRNA, and each has a specific functional location, whether in the cytoplasm, on cellular membranes, inside organelles, or in extracellular fluids. In this section we consider the movement and sorting of proteins from RER through vesicles of the endomembrane system, as well as the transport of proteins into and out of organelles.
17.7.1 Proteins Packaged in RER Are Made as Larger Precursor Proteins
All protein synthesis begins with the formation of an initiation complex and subsequent elongation cycles of peptide-bond formation and carboxyl-terminal amino acid addition. But proteins to be packaged for secretion or into lysosomes, peroxisomes, or other microbodies will complete translation elongation directly into the cisternae (enclosed spaces) of the RER. What traffic signal led some proteins to the RER and others elsewhere in the cytoplasm? A model system for studying secretory protein synthesis is mouse myeloma cells, cancerous lymphocytes of the immune system. Normal lymphocytes make immunoglobulin G (IgG) with light- and heavy-chain polypeptide subunits. IgGs are circulating antibodies. The myeloma cells make only IgG light chains, which are easily isolated from a cell culture medium.
In an early experiment, mouse myeloma cells were cultured under different conditions, and the secreted IgG light chain that was produced was analyzed. The results revealed that the secreted mouse light-chain IgG proteins that had been made in an in vitro translation system were in fact larger than proteins naturally secreted by the cells. The experimental protocol is illustrated in Figure 17.18.

In one part of this experiment, myeloma cells were grown in the presence of radioactive amino acids. The resulting radioactive IgG light-chain polypeptides secreted by the cells were isolated (follow the arrows across the top of Figure 17.18). In another part of the experiment, mRNA was isolated from a different batch of the myeloma cells and added to an in vitro translation system containing radioactive amino acids (follow the arrows down the left and across the bottom of the figure). The radioactive polypeptides made in vivo and in vitro were electrophoresed (follow the arrows to the gel) and the gels then autoradiographed. From the results at the lower right, you can see that the secreted polypeptides made in vivo had migrated faster on the gel than those translated in vitro. So, the cell-free translation product was indeed larger than the mature secreted polypeptide. To explain these results, Günter Blobel and colleagues extended the “Signal Hypothesis,” proposing that the signal was a short N-terminal signal peptide that directs a growing secretory polypeptide to the RER. They further proposed that the signal peptide is a temporary “traffic” signal, removed by an RERassociated enzyme as the polypeptide crossed the RER membrane into the cisternal space.
17.7.2 Testing the Signal Hypothesis for Packaging Secreted Proteins in RER
In the test of the Signal Hypothesis (which earned Blobel the 1999 Nobel Prize in Physiology or Medicine), isolates of RER membranes were included with mouse myeloma cell mRNA in cellfree protein-synthesis systems. This time, when secreted and cell-free synthesized IgG light chain polypeptides were electrophoresed and the gel was autoradiographed, both polypeptides were the same size as the mature, secreted polypeptides. Therefore, as predicted, RER contains processing activity—that is, a signal peptidase that removes the signal peptide! The steps of the signal hypothesis that emerged from the experiments of Blobel and his colleagues are illustrated in Figure 17.19.
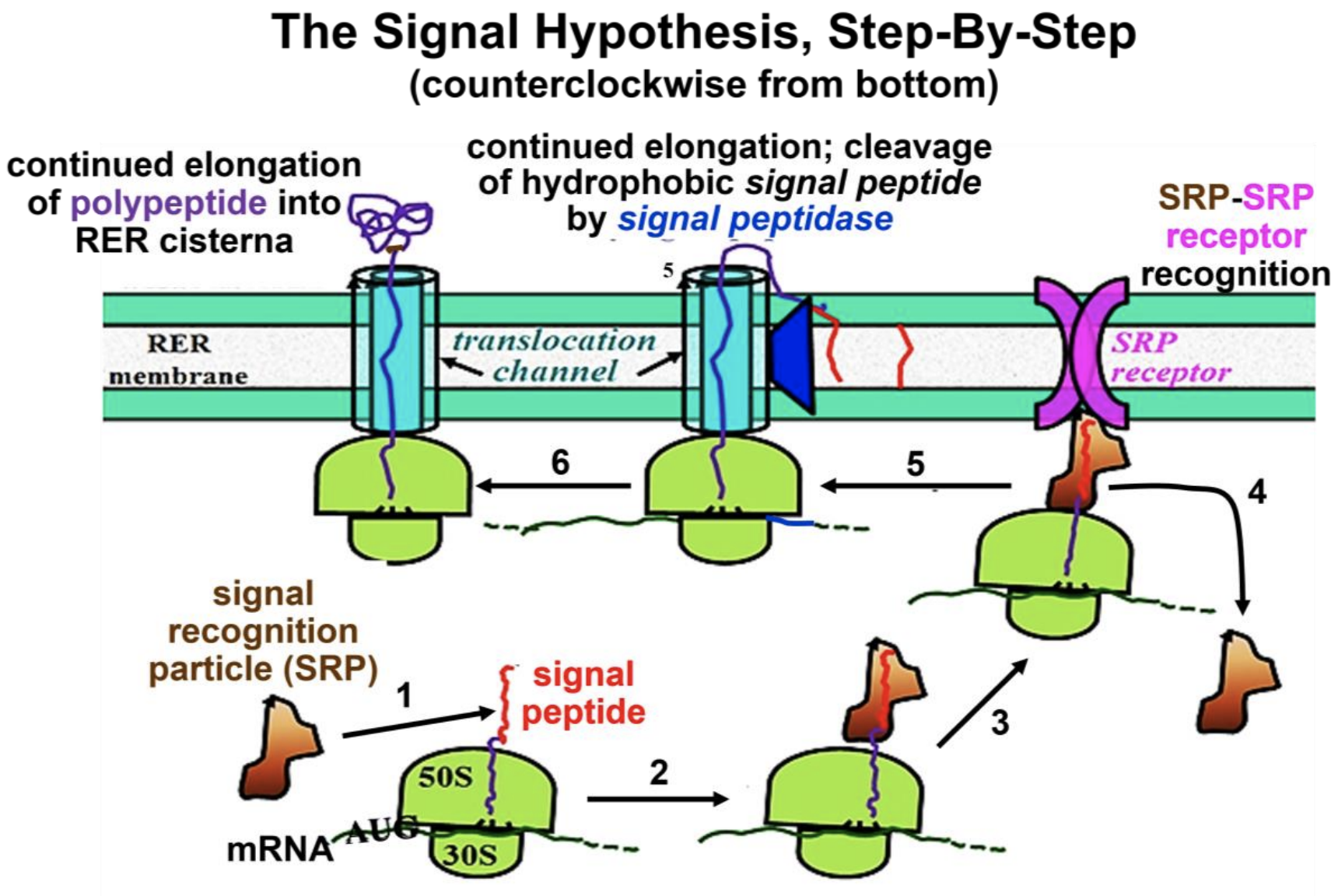
Recall that in translation, during polypeptide elongation, the growing polypeptide moves through and emerges from a channel or groove in the large ribosomal subunit. As the N terminal signal sequence (i.e., the signal peptide) of a secretory polypeptide emerges from this groove, it interacts with the RER membrane. Beginning at the lower left in Figure 17.19, the steps of protein packaging for the secretion process are these:
- An SRP (signal recognition particle) binds to the hydrophobic signal peptide.
- Elongation stops until the SRP-ribosome complex finds the RER membrane.
- The ribosome-SRP complex binds to an SRP receptor on the RER membrane.
- The SRP detaches from the growing polypeptide chain and is recycled.
- Translation elongation resumes through a translocation channel, and a signal peptidase in the RER membrane catalyzes cotranslational hydrolysis of the signal peptide, which remains embedded in the RER membrane.
- Elongation continues, and the growing polypeptide begins to fold in the RER.
305 Testing the Signal Hypothesis
306-2 Details of the Signal Hypothesis
Step 2 requires that the SRP find and bind to the signal peptide before the nascent polypeptide gets too long and starts folding into a 3D (tertiary) conformation that would hide the signal peptide. The ribosome itself may keep the signal peptide available by destabilizing electrostatic interactions that would otherwise lead to premature folding and an undoubtedly incorrect conformation. For more on ribosome involvement in protein folding, see Protein-Folding starts Near The Ribosome Making It.
Bacterial secretions include proteins involved in nutrient scavenging as well in cell wall synthesis, using a secretory mechanism like that of eukaryotes, with obvious differences in detail. Partially elongated signal peptides guide mRNA-bound ribosomes to the cytoplasmic side of the plasma membrane, where the ribosomes bind and then pass elongating proteins through the plasma membrane into the space between the cell membrane and wall. As the protein exits the cell, a bacterial signal peptidase (SPase) cleaves the signal peptide. Apparently, the basic mechanism for the secretion of proteins evolved early and is conserved in prokaryotes.
Early on, we knew that some antibiotics stop bacterial growth by disrupting the cell wall or by killing the cells outright. Others (e.g., arylomycins) disrupt plasma membrane SPase function, preventing proteins needed in the space between the cell wall and membrane from ever making it out of the cell. Once used against S. aureus, arylomycins are no longer effective because many Staphylococcus strains have become resistant to these antibiotics. Check out Bacterial SPase and Antibiotic Resistance to read about the mechanism of arylomycin resistance. As you may already know, S. aureus is now resistant to many antibiotics, and illness from untreatable infections has its own name, MRSA (Methicillin-Resistant S. Aureus—dig on your own to see more about methicillin resistance). While named for methicillin resistance, the MRSA acronym has also come to describe untreatable S. aureus infections in general.
Next we’ll look at how proteins get into organelles and membranes themselves.


