12.2: Gene Regulation in Prokaryotes- the Lactose (lac) Operon
- Page ID
- 88969
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Many prokaryotic genes are organized into operons: linked genes transcribed into a single mRNA that encodes two or more proteins. Operons usually encode proteins with related functions. Regulating the activity of an operon (rather than multiple single genes encoding single proteins) allows better coordination of the synthesis of several proteins at once. In E. coli, the regulated lac operon encodes three enzymes involved in the metabolism of lactose (an alternative nutrient to glucose).
Regulation of an operon (or of a single gene for that matter) may occur by repression or by induction. When a small metabolite in a cell binds to a regulatory repressor or inducer protein, the protein undergoes an allosteric change that allows it to bind to or to unbind from a regulatory DNA sequence.
We will see examples of such regulation in the lac and trp (tryptophan) operons. Regulation of the lac-operon genes is an example of gene repression as well as gene induction. We’ll see that trp-operon regulation is by gene repression. In both operons, changes in levels of intracellular metabolites reflect the metabolic status of the cell, eliciting appropriate changes in gene transcription. Let’s start with a look at lac-operon regulation.
216 Overview of Prokaryotic Gene Regulation
The mRNA transcribed from the lac operon is simultaneously translated into three enzymes, as shown in Figure 12.1.

12.2.1. Working Out Regulation of the lac Operon in E. coli
In the animal digestive tract (including ours), genes of the E. coli lac operon regulate the use of lactose as an alternative nutrient to glucose. Think “cheese” instead of “chocolate”! The operon consists of lacZ, lacY, and lacA genes, which are all structural genes.
By definition, structural genes encode proteins that participate in cell structure and metabolic function. As already noted, the lac operon is transcribed into an mRNA encoding the Z, Y, and A proteins. Let’s zoom in on a single lac operon in Figure 12.2.

The lacZ gene encodes β-galactosidase, the enzyme that breaks lactose, a disaccharide, into galactose and glucose. The lacY gene encodes lactose permease, a membrane protein that facilitates lactose entry into the cells. The role of the lacA gene (encoding a transacetylase) in lactose energy metabolism is not well understood. The lacI gene to the left of the lac Z gene is a regulatory gene (distinct from structural genes). Regulatory genes encode proteins that interact with the regulatory DNA sequences associated with a gene to control transcription. As we describe next, the operator sequence separating the lacI and lacZ genes is a transcription-regulatory DNA sequence.
The E. coli lac operon is usually silent (repressed) because these cells prefer glucose as an energy and carbon source. In the presence of sufficient glucose, a repressor protein (the lacI-gene product) is bound to the operator, blocking transcription of the lac operon. Even if lactose is available, cells will not be able to use it as an alternative energy and carbon source when glucose levels are adequate. However, when glucose levels drop, the lac operon is active, and the three enzyme products are translated. We will see how lac-operon transcription by both derepression and direct induction can lead to maximal transcription of the lac genes only when necessary (i.e., in the presence of lactose and absence of glucose). Let’s look at some of the classic experiments that led to our understanding of E. coli–gene regulation in general, and of the lac operon in particular.
In the late 1950s and early 1960s, François Jacob and Jacques Monod were studying the use of different sugars as carbon sources by E. coli. They knew that wild-type E. coli would not make the β-galactosidase, the β-galactoside permease, or the β-galactoside transacetylase proteins when grown on glucose. Of course, they also knew that the cells would switch to lactose for growth and reproduction if they were deprived of glucose! They then searched for and isolated different E. coli mutants that could not grow on lactose, even when there was no glucose in the growth medium.
Here are some of the mutants they studied:
- One mutant failed to make active β-galactosidase but did make permease.
- One mutant failed to make active permease but made normal amounts of β-galactosidase.
- Another mutant failed to make transacetylase but could still metabolize lactose in the absence of glucose. Hence the uncertainty of its role in lactose metabolism.
- Curiously, one mutant strain failed to make any of the three enzymes!
Since double mutants are very rare and triple mutants even rarer, François Jacob and Jacques Monod inferred that the activation of all three genes in the presence of lactose were controlled together in some way. In fact, it was this discovery that defined the operon as a set of genes transcribed as a single mRNA, whose expression could therefore be effectively coordinated. They later characterized the repressor protein produced by the lacI gene. F. Jacob, J. Monod, and A. Lwoff shared the Nobel Prize in Physiology or Medicine in 1965 for their work on bacterial gene regulation. We now know that there are several layers of lacoperon regulation. Negative and positive regulation of the operon depends on two regulatory proteins to control the rate of lactose metabolism.
12.2.2. Negative Regulation of the lac Operon by Lactose
Repression of lac-operon activity involves a repressor protein that must be removed for gene expression to occur. The repressor protein product of the I gene is always made and present in E. coli cells; expression of the I gene is constitutive, meaning it is unregulated. In the absence of lactose in the growth medium, the repressor protein binds tightly to the operator DNA. While RNA polymerase is bound to the promoter and ready to transcribe the operon, the presence of the repressor protein—which is bound to the operator sequence between the RNA polymerase and the Z-gene transcription start site—physically blocks its forward movement. Under these conditions, little or no transcript is made. Figure 12.3 shows the lac repressor protein in its repressed state.

If cells are grown in the presence of lactose, the lactose entering the cells is converted to allolactose. Allolactose in turn binds to the repressor sitting on the operator DNA to form a two-part complex, as shown in Figure 12.4.

The allosterically altered repressor dissociates from the operator, and RNA polymerase can resume transcribing the lac-operon genes (Figure 12.5).

12.2.3. Positive Regulation of the lac Operon: Induction by Catabolite Activation
The second control mechanism regulating lac-operon expression is mediated by CAP (cAMP-bound catabolite activator protein or cAMP receptor protein). When glucose is available, cellular levels of cAMP (cyclic AMP) are low in the cells, and CAP is in an inactive conformation. On the other hand, if glucose levels are low, cAMP levels rise and bind to the CAP, activating it. If lactose levels are also low, the cAMP-bound CAP will have no effect. If lactose is present and glucose levels are low, then allolactose binds the lac repressor, causing it to dissociate from the operator region. Under these conditions, the cAMP-bound CAP can bind to the operator in lieu of the repressor protein. In this case, rather than blocking RNA polymerase, the activated cAMP-bound CAP induces even more efficient lac-operon transcription. The result is synthesis of higher levels of lac enzymes, which facilitate efficient cellular use of lactose as an energy source alternative to glucose. Figure 12.6 shows maximal lac-operon activation in high lactose and low glucose.
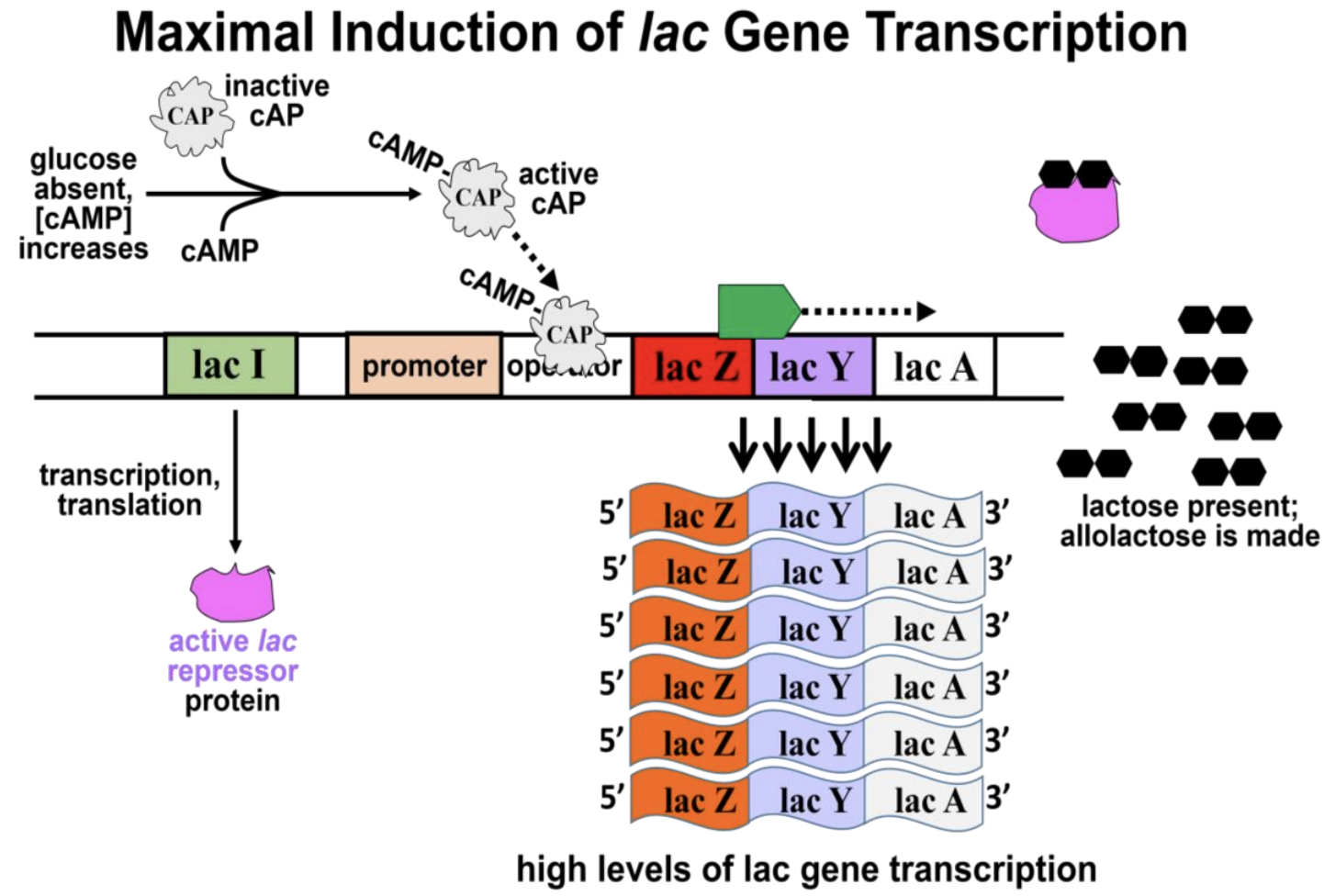
217 Regulation of the lac Operon
cAMP-bound-CAP is a transcription inducer. It forces DNA in the promoter-operator region to bend (Figure 12.7).

Binding of cAMP-CAP to the double helix loosens the H-bonds between the strands, making it easier for RNA polymerase to find and to bind the promoter and for transcription to begin.
In recent years, additional layers of lac-operon regulation have been uncovered. In one case, the ability of lac permease to transport lactose across the cell membrane is regulated. In another, additional operator sequences have been discovered to interact with a multimeric repressor to control lac-gene expression. We’ll consider inducer exclusion first.
12.2.4. lac-Operon Regulation by Inducer Exclusion
When glucose levels are high (even in the presence of lactose), phosphate is consumed to phosphorylate glycolytic intermediates. This keeps cytoplasmic phosphate levels low. Under these conditions, an unphosphorylated elongation factor (\(\rm EIIA^{Glc}\)) binds to the lactose permease in the cell membrane, preventing the enzyme from bringing lactose into the cell. The roles of phosphorylated and unphosphorylated \(\rm EIIA^{Glc}\) in regulating the lac operon are shown below in Figure 12.8.

High glucose levels block lactose entry into the cells, effectively preventing allolactose formation and derepression of the lac operon. Inducer exclusion is thus a logical way for the cells to handle an abundance of glucose, whether or not lactose is present. On the other hand, if glucose levels are low in the growth medium, phosphate concentrations in the cells rise sufficiently for a specific kinase to phosphorylate the \(\rm EIIA^{Glc}\). Then, phosphorylated \(\rm EIIA^{Glc}\) undergoes an allosteric change and dissociates from the lactose permease, making it active, so that more lactose can enter the cell. In other words, the inducer is not “excluded” under these conditions! The kinase that phosphorylates \(\rm EIIA^{Glc}\) is part of a phosphoenolpyruvate- (PEP-) dependent phosphotransferase system (PTS) cascade. When extracellular glucose levels are low, the cell activates the PTS system to bring whatever glucose is around into the cell. The last enzyme in the PTS phosphorylation cascade is the kinase that phosphorylates \(\rm EIIA^{Glc}\). Now activated, phosphorylated \(\rm EIIA^{Glc}\) dissociates from the lactose permease. The now-active permease can bring available lactose from the medium into the cell.
One can imagine several different mutations of the lac I gene that one can imagine affecting crucial functions of its repressor protein product. Hypothesize at least three such mutations and describe their effect on the lac repressor protein, and on lac operon gene expression.
12.2.5. Structure of the lac-Repressor Protein and Additional Operator Sequences
The lac repressor encoded by the I gene is a tetramer of identical subunits (Figure 12.9).

Each subunit contains a helix-turn-helix motif capable of binding to DNA. However, the operator DNA sequence downstream of the promoter in the operon consists of a pair of inverted repeats spaced apart in such a way that they can only interact with two of the repressor subunits, leaving the function of the other two subunits unknown…that is, until recently! Two more operator regions were recently characterized in the lac operon. Operator 2 (O2) is within the lac Z gene itself, and O3 is near the end but still within the lac I gene. Apart from their unusual location within actual genes, these operators, which interact with the remaining two repressor subunits, went undetected at first because mutations in the O2 or O3 regions individually do not contribute substantially to the effect of lactose in derepressing the lac operon. Only mutating both regions at the same time results in a substantial reduction in the binding of the repressor to the operon.


