9.2: DNA Replication
- Page ID
- 88948
As we’ve seen, single DNA strands have directionality, with a 5′ nucleotide phosphate end and a 3′ deoxyribose hydroxyl end. This is also true for circular bacterial chromosomes; you can see this if the circle is broken! Because the strands of the double helix are antiparallel, the 5′ end of one strand aligns with the 3′ end of the other at both ends of the double helix. In looking at the complementary pairing of bases of deoxynucleotides in the double helix, Watson and Crick quickly realized that the base sequence of one strand of DNA could serve as a template to make a new complementary strand. As we’ll see, this structure raised questions about DNA replication. Finding answers began with experiments that visualized the replication process.
9.2.1. Visualizing Replication and Replication Forks
Recall the phenomenon of bacterial conjugation showing that the bacterial chromosome (i.e., genome) is circular. In 1963, John Cairns confirmed this fact by direct visualization of bacterial DNA. He cultured E. coli cells for long periods on \({}^{3}\)H-thymidine (\({}^{3}\)H-T), a radioactive precursor to DNA, to make their entire cellular DNA radioactive. He then disrupted the cells gently to minimize damage to the DNA. The released DNA was allowed to settle and to adhere to membranes. Cairns placed a sensitive film over the membrane, allowed time for the radiation to expose the film and then developed the films (autoradiographs). He examined the autoradiographs in the electron microscope and saw tracks of silver grains (the same kind that create an image on film in old-fashioned photography) (drawn in Figure 9.1 below).
Cairns measured the length of the “silver” tracks, which most often consisted of three different closed loops, or circles (see the right-hand drawing in the figure). The circumferences of two of these circles were always equal, and their lengths, measured in nanometers, corresponded closely to a predicted DNA content of a single, non-dividing cell. Cairns therefore interpreted these images to be bacterial DNA in the process of replication.
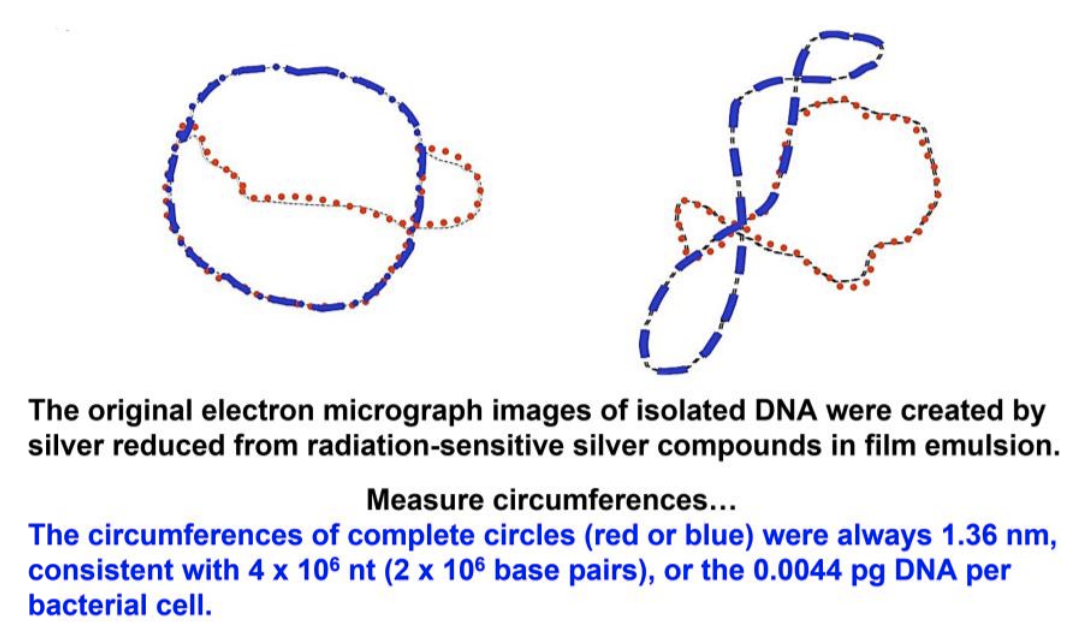
Cairns called his replicating chromosomes “theta images” because they resembled the Greek letter theta (\(\theta\)). From many \(\theta\) images, he arranged a sequence (shown in Figure 9.2) to illustrate his inference that replication starts at a single origin of replication on the bacterial chromosome, before proceeding around the circle at replication forks to completion.

Identify the three closed loops in on the left in Figure 9.1. Why does the third loop differ in circumference from the other two loops?
9.2.2. Visualizing Bidirectional Replication
David Prescott demonstrated bidirectional replication, which began at a replication origin but replicated DNA in opposite directions at not one but two replication forks (Figure 9.3).

176 Semiconservative Bidirectional Replication from Two RFs
A typical rate of DNA synthesis is about \(2 \times 10^6\) base pairs per hour, just about the size of the E. coli genome! A typical eukaryotic cell nucleus contains thousands of times more DNA than a bacterium but divides every fifteen to twenty hours. Even a small eukaryotic chromosome can contain hundreds or thousands of times as much DNA as a bacterium. Thus, eukaryotic cells can’t afford to replicate DNA at a bacterial rate! Eukaryotes solved this problem not by evolving faster replication biochemistry, but by synthesizing their DNA bidirectionally from multiple origins of replication, creating multiple replicons. These grow, joining other growing replicons along each linear chromosome, as suggested in Figure 9.4.

177 Multiple Replicons in Eukaryotes
Before we consider the biochemical events at replication forks in detail, let’s look at the role of DNA-polymerase enzymes in the process.


