5.6: Translation
- Page ID
- 1655
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Translation is the process by which information in mRNAs is used to direct the synthesis of proteins. As you have learned in introductory biology, in eukaryotic cells, this process is carried out in the cytoplasm of the cell, by large RNA-protein machines called ribosomes. Ribosomes contain ribosomal RNAs (rRNAs) and proteins. The proteins and rRNAs are organized into two subunits, a large and a small. The large subunit has an enzymatic activity, known as a peptidyl transferase, that makes the peptide bonds that join amino acids to make a polypeptide. The small and large subunits assemble on the mRNA at its 5’end to initiate translation. Ribosomes function by binding to mRNAs and holding them in a way that allows the amino acids encoded by the RNA to be joined sequentially to form a polypeptide.
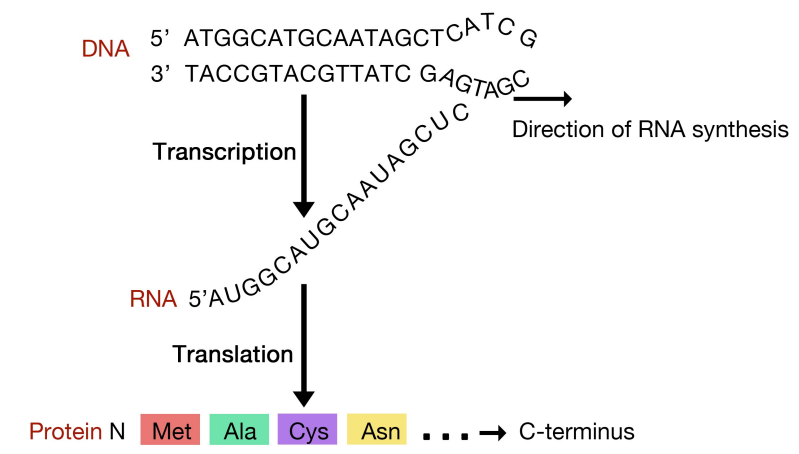
The sequence of an mRNA directly specifies the sequence of amino acids in the protein it encodes. Each amino acid in the protein is specified by a sequence of 3 bases called a codon in the mRNA. For example, the amino acid tryptophan is encoded by the sequence 5’UGG3’ on an mRNA. Given that there are 4 bases in RNA, the number of different 3-base combinations that are possible is \(4^3\), or 64. There are, however, only 20 amino acids that are used in building proteins. This discrepancy in the number of possible codons and the actual number of amino acids they specify is explained by the fact that the same amino acid may be specified by more than one codon. In fact, with the exception of the amino acids methionine and tryptophan, all the other amino acids are encoded by multiple codons. The figure above shows the codons that are used for each of the twenty amino acids.
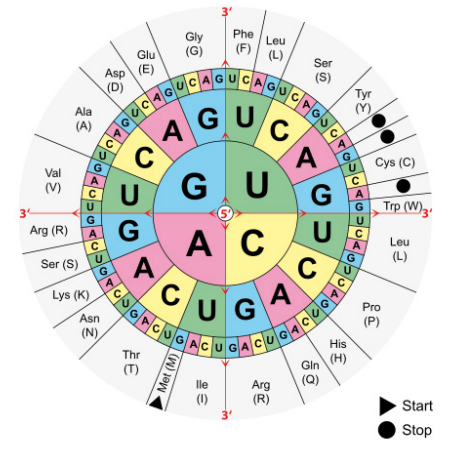
Three of the 64 codons are known as termination or stop codons and as their name suggests, indicate the end of a protein coding sequence. The codon for methionine, AUG, is used as the start, or initiation, codon.
This ingenious system is used to direct the assembly of a protein in the same way that you might string together colored beads in a particular order using instructions that used symbols like 111 for a red bead, followed by 222 for a green bead, 333 for yellow, and so on, till you came to 000, indicating that you should stop stringing beads.
While the ribosomes are literally the protein factories that join amino acids together using the instructions in mRNAs, another class of RNA molecules, the transfer RNAs (tRNAs) are also needed for translation. Transfer RNAs (see figure, left) are small RNA molecules, about 75-80 nucleotides long, that function to 'interpret' the instructions in the mRNA during protein synthesis. In terms of the bead analogy above, someone, or something, has to be able to bring a red bead in when the instructions indicate 111, and a green bead when the instructions say 222. Unlike a human, who can choose a red bead when 111 is present in the instructions, neither ribosomes nor tRNAs can think. The system, therefore, relies, like so many processes in cells, solely on molecular recognition.
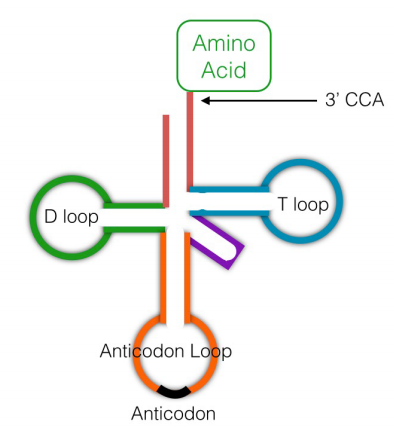
A given transfer RNA is specific for a particular amino acid. It is linked covalently to this amino acid at its 3' end by an enzyme called aminoacyl tRNA synthetase. There is an aminoacyl tRNA synthetase specific for each amino acid. A tRNA with an amino acid attached to it is said to be charged. Another region of the tRNA has a sequence of 3 bases, the anticodon, that is complementary to the codon for the amino acid it is carrying. When the tRNA encounters the codon for its amino acid on the messenger RNA, the anticodon will base-pair with the codon, and the amino acid attached to it will be brought in to the ribosome to be added on to the growing protein chain.
With an idea of the various components necessary for translation we can now take a look at the process of protein synthesis. The main steps in the process are similar in prokaryotes and eukaryotes. As we already noted, ribosomes bind to mRNAs and facilitate the interaction between the codons in the mRNA and the anticodons on charged tRNAs.
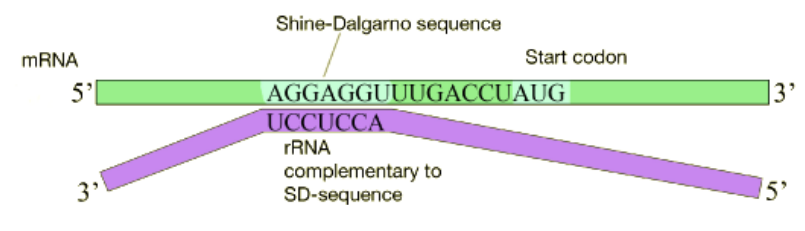
In bacterial cells, translation is coupled with transcription and begins even before the mRNA has been completely synthesized. How does the ribosome recognize and bind to the mRNA. Many bacterial mRNAs carry a short purine-rich sequence known as the Shine-Dalgarno site upstream of the AUG start codon, as shown in the figure below. This sequence is recognized and bound by a complementary sequence in the 16S rRNA that is part of the small ribosomal subunit as shown above. Because the Shine-Dalgarno site serves to recruit and bind the ribosome, it is also referred to as the Ribosome Binding Site or RBS.

A variation of this process of ribosome assembly operates in eukaryotic cells. We already know that in eukaryotic cells, processed mRNAs are sent from the nucleus to the cytoplasm.
The small and large subunits of ribosomes, each composed of characteristic rRNAs and proteins are found in the cytoplasm and assemble on mRNAs to form complete ribosomes that carry out translation.
Protein synthesis in eukaryotes starts with the binding of the small subunit of the ribosome to the 5' end of the mRNA. The assembly of the translation machinery begins with the binding of the small ribosomal subunit to the 7-methyl guanosine cap on the 5'end of an mRNA. Meanwhile, the initiator tRNA pairs with the start codon. (Recall that the start codon is AUG, and codes for methionine. The initiator tRNA carries the amino acid methionine). The large subunit of the ribosome then joins the complex, which is now ready to start protein synthesis.
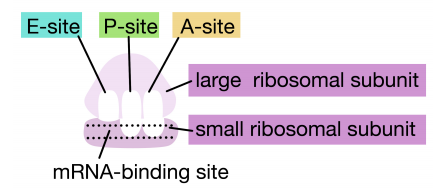
Ribosomes have two sites for binding charged tRNAs, each of which is positioned to make two adjacent codons on the mRNA available for binding by tRNAs. The initiation codon occupies the first of these ribosomal sites, the P-site. The anticodon complementary to this is on the initiator tRNA, which brings in the first amino acid of the protein. This initial phase of translation is called initiation and requires the help of protein factors called initiation factors.
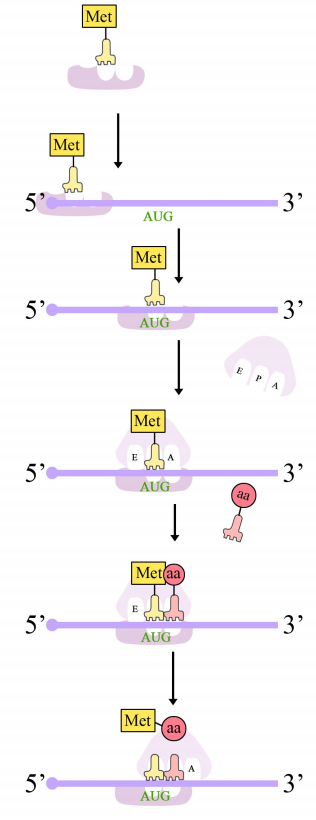
The second codon of the mRNA is positioned adjacent to the second site on the ribosome, the A site. This is where the tRNA carrying the amino acid specified by the second codon binds. The binding of aminoacyl tRNA to the A-site is mediated by proteins called elongation factors and requires the input of energy. Once the appropriate charged tRNAS have "docked" on the codons by base-pairing between the anticodon on the tRNA and the codon on the mRNA, the ribosome joins the amino acids carried by the two tRNAs by making a peptide bond (see figure at right).
Interestingly, the formation of the peptide bond is catalyzed by a catalytic RNA (the 23S rRNA in prokaryotes) rather than by a protein enzyme.
This and subsequent steps in the synthesis of the polypeptide are called the elongation phase of translation. Once the first two amino acids are linked , the first tRNA dissociates, and moves out of the P-site and into the E, or Exit site. The second tRNA then moves into the P-site, vacating the A-site for the tRNA corresponding to the next codon.
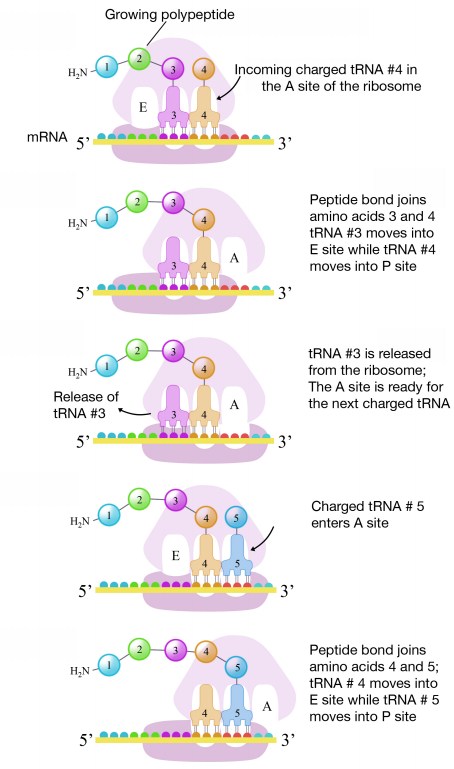
The process repeats till the stop codon is in the A-site. At this point, a release factor binds at the A-site, adds a water molecule to the polypeptide at the P-site, and releases the completed polypeptide from the ribosome, which itself, then dissociates into subunits.
As described in Chapter 3, polypeptides made in this way are then folded into their three dimensional shapes, post-translationally modified and delivered to the appropriate cellular compartments to carry out their functions.


