2017_SS1_Lecture_05
- Page ID
- 9957
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Introduction to bacterial and archaeal diversity
Perhaps bacteria may tentatively be regarded as biochemical experiments; owing to their relatively small size and rapid growth, variations must arise much more frequently than in more differentiated forms of life, and they can in addition afford to occupy more precarious positions in natural economy than larger organisms with more exacting requirements.
Marjory Stephenson, in Bacterial Metabolism, (1930)
Prokaryotes are single-celled organisms with neither a membrane-bound nucleus nor other lipid membrane-bound organelles. They are composed of two phylogenetically distinct groups of organisms: Bacteria and Archaea. In recent years, the term prokaryote has fallen out of favor for many microbiologists. The reason is that while bacteria and archaea share many morphological characteristics, they nevertheless, represent evolutionarily distinct domains of life. The figure below shows a simple phylogenetic tree with the three main domains of life: Bacteria, Archaea, and Eukarya. This means that the use of the term prokaryote should not be used with the intention to group the bacteria and archaea on the basis of shared evolutionary history. It is, however, convenient to use the term "prokaryote" when describing the groups of organisms that share the common morphological characteristics (i.e. no nucleus) and some of your instructors will likely do so. When you hear or use the term "prokaryote", therefore, make sure that it is not being used to or implying that the bacteria and archaea are part of the same phylogenetic group. Rather make sure that the use of the term "prokaryote" is limited to describing the common physical characteristics of these two microbial groups.

Figure 1. Although bacteria and archaea are both described as prokaryotes, they have been placed in separate domains of life. An ancestor of modern archaea is believed to have given rise to Eukarya, the third domain of life. Archaeal and bacterial phyla are shown; the exact evolutionary relationship between these phyla is still open to debate.
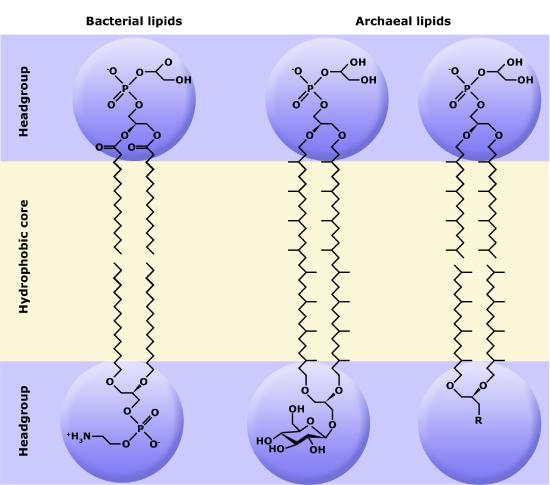
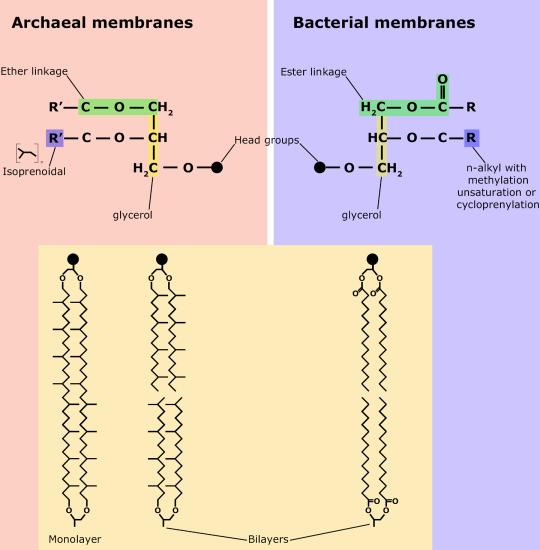
Although bacteria and archaea share many morphological, structural, and metabolic attributes, there are numerous differences between the organisms in these two clades. The most notable differences are in the chemical structure and compositions of membrane lipids, the chemical composition of the cell wall, and the makeup of the information processing machinery (e.g., replication, DNA repair, and transcription).
Bacterial and archaeal diversity
Bacteria and archaea were on Earth long before multicellular life appeared. They are ubiquitous and have highly diverse metabolic activities. This diversity allows different species within clades to inhabit every imaginable surface where there is sufficient moisture. For example, some estimates suggest that in the typical human body, bacterial cells outnumber human body cells by about ten to one. Indeed, bacteria and archaea comprise the majority of living things in all ecosystems. Certain bacterial and archaeal species can thrive in environments that are inhospitable for most other life. Bacteria and archaea, along with microbial eukaryotes, are also critical for recycling the nutrients essential for creating new biomolecules. They also drive the evolution of new ecosystems (natural or man-made).
The first inhabitants of Earth
The Earth and its moon are thought to be about 4.54 billion years old. This estimate is based on evidence from radiometric dating of meteorite material, together with other substrate material from Earth and the moon. Early Earth had a very different atmosphere (contained less molecular oxygen) than it does today and was subjected to strong radiation; thus, the first organisms would have flourished in areas where they were more protected, such as in ocean depths or beneath the Earth's surface. During this time period, strong volcanic activity was common on Earth, so it is likely that these first organisms were adapted to very high temperatures. Early Earth was also bombarded with mutagenic radiation from the sun. The first organisms, therefore, needed to be able to withstand all these harsh conditions.
So, when and where did life begin? What were the conditions on Earth when life began? What did LUCA (the Last Universal Common Ancestor), the predecessor to bacteria and archaea look like? While we don't know exactly when and how life arose and what it looked like when it did, we do have a number of hypotheses based on various biological and geological data that we briefly describe below.
The ancient atmosphere
Evidence indicates that during the first two billion years of Earth’s existence, the atmosphere was anoxic, meaning that there was no molecular oxygen. Therefore, only those organisms that can grow without oxygen—anaerobic organisms—were able to live. Autotrophic organisms that convert solar energy into chemical energy are called phototrophs, and they appeared within one billion years of the Earth's formation. Then, cyanobacteria, also known as blue-green algae, evolved from these simple phototrophs one billion years later. Cyanobacteria began oxygenating the atmosphere. Increased atmospheric oxygen allowed the development of more efficient O2-utilizing catabolic pathways. It also opened up the land to increased colonization, because some O2 is converted into O3 (ozone), and ozone effectively absorbs the ultraviolet light that would otherwise cause lethal mutations in DNA. Ultimately, the increase in O2 concentrations allowed the evolution of other life forms.
Note:
The evolution of bacteria and archaea:
How do scientists answer questions about the evolution of bacteria and archaea? Unlike with animals, artifacts in the fossil record of bacteria and archaea offer very little information. Fossils of ancient bacteria and archaea look like tiny bubbles in rock. Some scientists turn to comparative genetics which, as its name suggests, is a domain of biology that makes quantitative comparisons of the genetic information between two or more species. A core assumption in the field of comparative genetics is that the more recently two species have diverged, the more similar their genetic information will be. Conversely, species that diverged long ago will have more genes that are dissimilar. Therefore, by comparing genetic sequences between organisms can shed light on their evolutionary relationships and allow scientists to create models of what the genetic makeup of the ancestors of the organisms being compared might have looked like.
Scientists at the NASA Astrobiology Institute and at the European Molecular Biology Laboratory collaborated to analyze the molecular evolution of 32 specific proteins common to 72 species of bacteria. The model they derived from their data indicates that three important groups of bacteria—Actinobacteria, Deinococcus, and Cyanobacteria (which the authors call Terrabacteria)—were likely the first to colonize land. Organisms in the genus Deinococcus are bacteria that tend to be highly resistant to ionizing radiation. Cyanobacteria are photosynthesizers, while Actinobacteria are a group of very common bacteria that include species important in decomposition of organic wastes.
The timelines of species divergence suggest that bacteria (members of the domain Bacteria) diverged from common ancestral species between 2.5 and 3.2 billion years ago, whereas archaea diverged earlier: between 3.1 and 4.1 billion years ago. Eukarya diverged off the Archaean line later. Furthermore, there were bacteria able to grow in the anoxic environment that existed prior to the advent of cyanobacteria (about 2.6 billion years ago). These bacteria needed to be resistance to drying and to possess compounds that protect the organism from radiation. It has been proposed that the emergence of cyanobacteria with its ability to conduct photosynthesis and produce oxygen was a key event in the evolution of life on Earth.
Microbial mats
Microbial mats (large biofilms) may be representative of the earliest visible structure formed by life on Earth; there is fossil evidence of their presence starting about 3.5 billion years ago. A microbial mat is a multi-layered sheet of microbes composed mostly of bacteria but that may also include archaea. Microbial mats are a few centimeters thick, and they typically grow at the interface between two materials, mostly on moist surfaces. Organisms in a microbial mat are held together by a glue-like, sticky substance that they secrete, forming an extracellular matrix. The species within the mat carry out different metabolic activities depending on their environment. As a result, microbial mats have been identified that have different textures and colors reflecting the mat composition and the metabolic activities conducted by the microorganisms that make up the mat.
The first microbial mats likely harvested energy through redox reactions (discussed elsewhere) from chemicals found near hydrothermal vents. A hydrothermal vent is a breakage or fissure in the Earth’s surface that releases geothermally heated water. With the evolution of photosynthesis about 3 billion years ago, some organisms in microbial mats came to use a more widely available energy source—sunlight—whereas others depended on chemicals from hydrothermal vents for energy and food.

Figure 2. (a) This microbial mat, about one meter in diameter, grows over a hydrothermal vent in the Pacific Ocean in a region known as the “Pacific Ring of Fire.” Chimneys, such as the one indicated by the arrow, allow gases to escape. (b) In this micrograph, bacteria within a mat are visualized using fluorescence microscopy. (credit a: modification of work by Dr. Bob Embley, NOAA PMEL, Chief Scientist; credit b: modification of work by Ricardo Murga, Rodney Donlan, CDC; scale-bar data from Matt Russell)
Stromatolites
A stromatolite is a sedimentary structure formed when minerals precipitate out of water due to the metabolic activity of organisms in a microbial mat. Stromatolites form layered rocks made of carbonate or silicate. Although most stromatolites are artifacts from the past, there are places on Earth where stromatolites are still forming. For example, growing stromatolites have been found in the Anza-Borrego Desert State Park in San Diego County, California.

Figure 3. (a) These living stromatolites are located in Shark Bay, Australia. (b) These fossilized stromatolites, found in Glacier National Park, Montana, are nearly 1.5 billion years old. (credit a: Robert Young; credit b: P. Carrara, NPS).
Bacteria and archaea are adaptable: life in moderate and extreme environments
Some organisms have developed strategies that allow them to survive harsh conditions. Bacteria and archaea thrive in a vast array of environments: some grow in conditions that would seem very normal to us, whereas others are able to thrive and grow under conditions that would kill a plant or an animal. Almost all bacteria and archaea have some form of a cell wall, a protective structure that allows them to survive in both hyper- and hypo-osmotic conditions. Some soil bacteria are able to form endospores that resist heat and drought, thereby allowing the organism to survive until more favorable conditions recur. These adaptations, along with others, allow bacteria to be the most abundant life forms in all terrestrial and aquatic ecosystems.
Some bacteria and archaea are adapted to grow under extreme conditions and are called extremophiles, meaning “lovers of extremes.” Extremophiles have been found in all kinds of environments, such as in the depths of the oceans and the earth; in hot springs, the Artic, and the Antarctic; in very dry places; in harsh chemical environments; and in high-radiation environments, just to mention a few. These organisms help to give us a better understanding of the diversity of life and open up the possibility of finding microbial species that may lead to the discovery of new therapeutic drugs or have industrial applications. Because they have specialized adaptations that allow them to live in extreme conditions, many extremophiles cannot survive in moderate environments. There are many different groups of extremophiles. They are categorized based on the conditions in which they grow best, and several habitats are extreme in multiple ways. For example, a soda lake is both salty and alkaline, so organisms that live in a soda lake must be both alkaliphiles and halophiles. Other extremophiles, like radioresistant organisms, do not prefer an extreme environment (in this case, one with high levels of radiation) but have adapted to survive in it.
Table 1. This table lists some extremophiles and their preferred conditions.
| Extremophile Type | Conditions for Optimal Growth |
| Acidophiles | pH 3 or below |
| Alkaliphiles | pH 9 or above |
| Thermophiles | Temperature of 60–80 °C (140–176 °F) |
| Hyperthermophiles | Temperature of 80–122 °C (176–250 °F) |
| Psychrophiles | Temperature of -15 °C (5 °F) or lower |
| Halophiles | Salt concentration of at least 0.2 M |
| Osmophiles | High sugar concentration |

Figure 4. Deinococcus radiodurans, visualized in this false-color transmission electron micrograph, is a bacterium that can tolerate very high doses of ionizing radiation. It has developed DNA repair mechanisms that allow it to reconstruct its chromosome even if it has been broken into hundreds of pieces by radiation or heat. (credit: modification of work by Michael Daly; scale-bar data from Matt Russell)
Footnotes
1. Battistuzzi, FU, Feijao, A, and Hedges, SB. A genomic timescale of prokaryote evolution: Insights into the origin of methanogenesis, phototrophy, and the colonization of land. BioMed Central: Evolutionary Biology 4 (2004): 44, doi:10.1186/1471-2148-4-44.
Cellular structure of bacteria and archaea
In this section, we discuss the basic structural features of both bacteria and archaea. There are many structural, morphological, and physiological similarities between bacteria and archaea but the key feature is that both bacteria and archaea lack a membrane-bound nucleus and membrane-bound organelles, which are hallmarks of eukaryotes. As discussed earlier, many of these microbes inhabit diverse ecological niches and carry most the wide range of biochemical and metabolic processes on Earth.
While Bacteria and Archaea represent separate domains of life, morphologically they share a number of structural features. As a result, they face similar problems, such as the transport of nutrients into the cell, the removal of waste material from the cell, and the need to respond to rapid changes in the local environment. In this section, we will focus on how their common feature allows them to thrive in various environments and simultaneously puts constraints on them that can be used to explain how these cells function. One of the biggest constraints is related to cell size.
Although bacteria and archaea come in a variety of shapes, the most common three shapes are: cocci (spherical), bacilli (rod-shaped), and spirilli (spiral-shaped) (figure below). Both bacteria and archaea are generally small compared to typical eukaryotes. For example, most bacteria tend to be on the order of 0.2 to 1.0 µm (micrometers) in diameter and 1-10 µm in length. However, there are exceptions. Epulopiscium fishelsoni is a bacillus-shaped bacterium that is typically 80 µm in diameter and 200-600 µm long. Thiomargarita namibiensis is a spherical bacterium between 100 and 750 µm in diameter and is visible to the naked eye. For comparison, a typical human neutrophil is approximately 50 µm in diameter.
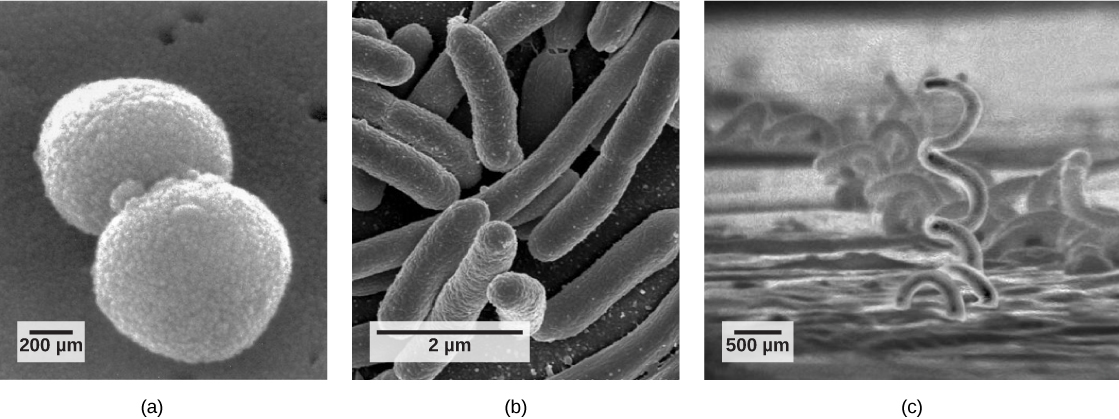
Figure 1. This figure shows the three most common shapes of bacteria and archaea: (a) cocci (spherical), (b) bacilli (rod-shaped), and (c) spirilli (spiral-shaped).
A thought question:
One question that comes to mind is why are bacteria and archaea typically so small? What are the functional and evolutionary advantages and disadvantages of being small? Are there physical constraints limiting size? If so, how could bacteria such as Epulopiscium fishelsoni and Thiomargarita namibiensis overcome these constraints? Think of possible explanations or hypotheses that might answer these questions. We'll explore and develop an understanding of these questions in more detail below and in class.
The bacterial and archaeal cell: common structures
Introduction to the basic cell structure
Bacteria and archaea are unicellular organisms, which lack internal membrane-bound structures that are disconnected from the plasma membrane, the phospholipid membrane that defines the boundary between the inside and outside of the cell. Rather, in bacteria and archaea, the cytoplasmic membrane contains all membrane-bound reactions, including those related to the electron transport chain, ATP synthase, and photosynthesis. By definition, these cells lack a nucleus. Instead, their genetic material is located in a self-defined area of the cell called the nucleoid. The bacterial and archaeal chromosome is often a single covalently closed circular double-stranded DNA molecule. However, some bacteria have linear chromosomes, and some bacteria and archaea have more than one chromosome or small non-essential circular replicating elements of DNA called plasmids. Besides the nucleoid, the next common feature is the cytoplasm (or cytosol), the "aqueous," jelly-like region encompassing the internal portion of the cell. The cytoplasm is where the soluble (non-membrane-associated) reactions occur and contains the ribosomes, the protein-RNA complex where proteins are synthesized. Finally, many bacteria and archaea also have cell walls, the rigid structural feature surrounding the plasma membrane that helps provide protection and constrain the cell shape. You should learn to create a simple sketch of a general bacterial or archaeal cell from memory.

Figure 2. The features of a typical prokaryotic cell are shown.
Constraints on the bacterial and archaeal cell
One common, almost universal, feature of bacteria and archaea is that they are small, microscopic to be exact. Even the two examples given as exceptions, Epulopiscium fishelsoni and Thiomargarita namibiensis, still face the basic constraints all bacteria and archaea face; they simply found unique strategies around the problem. So what is the largest constraint when it comes to dealing with the size of bacteria and archaea? Think about what the cell must do to survive.
Some basic requirements for survival
So what do cells have to do to survive? They need to transform energy into a usable form. This involves making ATP, maintaining an energized membrane, and maintaining productive NAD+/NADH2 ratios. Cells also need to be able to synthesize key macromolecules (proteins, lipids, polysaccharides, etc.) and other cellular structural components. To do this, they need to be able to either make the core, key precursors for more complex molecules or get them from the environment.
Diffusion and its importance to bacteria and archaea
Movement by diffusion is passive and proceeds down the concentration gradient. For compounds to move from the outside to the inside of the cell, the compound must be able to cross the phospholipid bilayer. If the concentration of a substance is lower inside the cell than outside and it has chemical properties that allow it to move across the cell membrane, that compound will energetically tend to move into the cell. While the "real" story is a bit more complex and will be discussed in more detail later, diffusion is one of the mechanisms bacteria and archaea use to aid in the transport of metabolites.
Diffusion can also be used to get rid of some waste materials. As waste products accumulate inside the cell, their concentration rises compared to that of the outside environment, and the waste product can leave the cell. Movement within the cell works the same way: compounds will move down their concentration gradient, away from where they are synthesized to places where their concentration is low and therefore may be needed. Diffusion is a random process—the ability of two different compounds or reactants for chemical reactions to interact becomes a meeting of chance. Therefore, in small, confined spaces, random interactions or collisions can occur more frequently than they can in large spaces.
The ability of a compound to diffuse depends on the viscosity of the solvent. For example, it is a lot easier for you to move around in air than in water (think about moving around underwater in a pool). Likewise, it is easier for you to swim in a pool of water than in a pool filled with peanut butter. If you put a drop of food coloring into a glass of water, it quickly diffuses until the entire glass has changed color. Now what do you think would happen if you put that same drop of food coloring into a glass of corn syrup (very viscous and sticky)? It will take a lot longer for the glass of corn syrup to change color.
The relevance of these examples is to note that the cytoplasm tends to be very viscous. It contains many proteins, metabolites, small molecules, etc. and has a viscosity more like corn syrup than water. So, diffusion in cells is slower and more limited than you might have originally expected. Therefore, if cells rely solely on diffusion to move compounds around, what do you think happens to the efficiency of these processes as cells increase in size and their internal volumes get bigger? Is there a potential problem to getting big that is related to the process of diffusion?
So how do cells get bigger?
As you've likely concluded from the discussion above, with cells that rely on diffusion to move things around the cell—like bacteria and archaea—size does matter. So how do you suppose Epulopiscium fishelsoni and Thiomargarita namibiensis got so big? Take a look at these links, and see what these bacteria look like morphologically and structurally: Epulopiscium fishelsoni and Thiomargarita namibiensis.
Based on what we have just discussed, in order for cells to get bigger, that is, for their volume to increase, intracellular transport must somehow become independent of diffusion. One of the great evolutionary leaps was the ability of cells (eukaryotic cells) to transport compounds and materials intracellularly, independent of diffusion. Compartmentalization also provided a way to localize processes to smaller organelles, which overcame another problem caused by the large size. Compartmentalization and the complex intracellular transport systems have allowed eukaryotic cells to become very large in comparison to the diffusion-limited bacterial and archaeal cells. We'll discuss specific solutions to these challenges in the following sections.
Membranes
Plasma membranes enclose and define the borders between the inside and the outside of cells. They are typically composed of dynamic bilayers of phospholipids into which various other lipid soluble molecules and proteins have also been embedded. These bilayers are asymmetric—the outer leaf being different than the inner leaf in lipid composition and in the proteins and carbohydrates that are displayed to either the inside or outside of the cell. Various factors influence the fluidity, permeability, and various other physical properties of the membrane. These include the temperature, the configuration of the fatty acid tails (some kinked by double bonds), the presence of sterols (i.e., cholesterol) embedded in the membrane, and the mosaic nature of the proteins embedded within it. The cell membrane has selectivity; it allows only some substances through while excluding others. In addition, the plasma membrane must, in some cases, be flexible enough to allow certain cells, such as amoebae, to change shape and direction as they move through the environment, hunting smaller, single-celled organisms.
Amoebae Hunting Video
Cellular membranes
A subgoal in our "build-a-cell" design challenge is to create a boundary that separates the "inside" of the cell from the environment "outside". This boundary needs to serve multiple functions that include:
- Act as a barrier by blocking some compounds from moving in and out of the cell.
- Be selectively permeable in order to transport specific compounds into and out of the cell.
- Receive, sense, and transmit signals from the environment to inside of the cell.
- Project "self" to others by communicating identity to other nearby cells.

Figure 1. The diameter of a typical balloon is 25cm and the thickness of the plastic of the balloon of around 0.25mm. This is a 1000X difference. A typical eukaryotic cell will have a cell diameter of about 50µm and a cell membrane thickness of 5nm. This is a 10,000X difference.
Note: possible discussion
The ratio of membrane thickness compared to the size of an average eukaryotic cell is much greater compared to that of a balloon stretched with air. To think that the boundary between life and nonlife is so small, and seemingly fragile, more so than a balloon, suggests that structurally the membrane must be relatively stable. Discuss why cellular membranes are stable. You will need to pull from information we have already covered in this class.
Fluid mosaic model
The existence of the plasma membrane was identified in the 1890s, and its chemical components were identified in 1915. The principal components identified at that time were lipids and proteins. The first widely accepted model of the plasma membrane’s structure was proposed in 1935 by Hugh Davson and James Danielli; it was based on the “railroad track” appearance of the plasma membrane in early electron micrographs. They theorized that the structure of the plasma membrane resembles a sandwich, with protein being analogous to the bread, and lipids being analogous to the filling. In the 1950s, advances in microscopy, notably transmission electron microscopy (TEM), allowed researchers to see that the core of the plasma membrane consisted of a double, rather than a single, layer. A new model that better explains both the microscopic observations and the function of that plasma membrane was proposed by S.J. Singer and Garth L. Nicolson in 1972.
The explanation proposed by Singer and Nicolson is called the fluid mosaic model. The model has evolved somewhat over time, but it still best accounts for the structure and functions of the plasma membrane as we now understand them. The fluid mosaic model describes the structure of the plasma membrane as a mosaic of components—including phospholipids, cholesterol, proteins, and carbohydrates—that gives the membrane a fluid character. Plasma membranes range from 5 to 10 nm in thickness. For comparison, human red blood cells, visible via light microscopy, are approximately 8 µm wide, or approximately 1,000 times wider than a plasma membrane.

Figure 2. The fluid mosaic model of the plasma membrane describes the plasma membrane as a fluid combination of phospholipids, cholesterol, and proteins. Carbohydrates attached to lipids (glycolipids) and to proteins (glycoproteins) extend from the outward-facing surface of the membrane.
The principal components of a plasma membrane are lipids (phospholipids and cholesterol), proteins, and carbohydrates. The proportions of proteins, lipids, and carbohydrates in the plasma membrane vary with organism and cell type, but for a typical human cell, proteins account for about 50 percent of the composition by mass, lipids (of all types) account for about 40 percent of the composition by mass, and carbohydrates account for the remaining 10 percent of the composition by mass. However, the concentration of proteins and lipids varies with different cell membranes. For example, myelin, an outgrowth of the membrane of specialized cells, insulates the axons of the peripheral nerves, contains only 18 percent protein and 76 percent lipid. The mitochondrial inner membrane contains 76 percent protein and only 24 percent lipid. The plasma membrane of human red blood cells is 30 percent lipid. Carbohydrates are present only on the exterior surface of the plasma membrane and are attached to proteins, forming glycoproteins, or to lipids, forming glycolipids.
Phospholipids
Phospholipids are major constituents of the cell membrane, the outermost layer of cells. Like fats, they are composed of fatty acid chains attached to a polar head group. Specifically, there are two fatty acid tails and a phosphate group as the polar head group. The phospholipid is an amphipathic molecule, meaning it has a hydrophobic part and a hydrophilic part. The fatty acid chains are hydrophobic and cannot interact with water, whereas the phosphate-containing head group is hydrophilic and interacts with water.
Note
Make sure to note in Figure 3 that the phosphate group has an R group linked to one of the oxygen atoms. R is a variable commonly used in these types of diagrams to indicate that some other atom or molecule is bound at that position. That part of the molecule can be different in different phospholipids—and will impart some different chemistry to the whole molecule. At the moment, however, you are responsible for being able to recognize this type of molecule (no matter what the R group is) because of the common core elements—the glycerol backbone, the phosphate group, and the two hydrocarbon tails.

Figure 3. A phospholipid is a molecule with two fatty acids and a modified phosphate group attached to a glycerol backbone. The phosphate may be modified by the addition of charged or polar chemical groups. Several chemical R groups may modify the phosphate. Choline, serine, and ethanolamine are shown here. These attach to the phosphate group at the position labeled R via their hydroxyl groups.
Attribution: Marc T. Facciotti (own work)
A phospholipid bilayer forms as the basic structure of the cell membrane. The fatty acid tails of phospholipids face inside, away from water, whereas the phosphate group faces outside, hydrogen bonding with water. Phospholipids are responsible for the dynamic nature of the plasma membrane.

Figure 4. In the presence of water, some phospholipids will spontaneously arrange themselves into a micelle. The lipids will be arranged such that their polar groups will be on the outside of the micelle, and the nonpolar tails will be on the inside. A lipid bilayer can also form, a two layered sheet only a few nanometers thick. The lipid bilayer consists of two layers of phospholipids organized in a way that all the hydrophobic tails align side by side in the center of the bilayer and are surrounded by the hydrophilic head groups.
Source: Created by Erin Easlon (own work)
Note: possible discussion
Above it says that if you were to take some pure phospholipids and drop them into water that some if it would spontaneously (on its own) form into micelles. This sounds a lot like something that could be described by an energy story. Go back to the energy story rubric and try to start creating an energy story for this process—I expect that the steps involving the description of energy might be difficult at this point (we'll come back to that later) but you should be able to do at least the first three steps. You can constructively critique (politely) each other's work to create an optimized story.
Note: possible discussion
Note that the phospholipid depicted above has an R group linked to the phosphate group. Recall that this designation is generic—these can be different than the R groups on amino acids. What might be a benefit/purpose of "functionalizing" or "decorating" different lipids with different R groups? Think of the functional requirements for membranes stipulated above.
Membrane proteins
Proteins make up the second major component of plasma membranes. Integral membrane proteins are, as their name suggests, integrated completely into the membrane structure, and their hydrophobic membrane-spanning regions interact with the hydrophobic region of the the phospholipid bilayer. Single-pass integral membrane proteins usually have a hydrophobic transmembrane segment that consists of 20–25 amino acids. Some span only part of the membrane—associating with a single layer—while others stretch from one side of the membrane to the other, and are exposed on either side. This type of protein has a hydrophilic region or regions, and one or several mildly hydrophobic regions. This arrangement of regions of the protein tends to orient the protein alongside the phospholipids, with the hydrophobic region of the protein adjacent to the tails of the phospholipids and the hydrophilic region or regions of the protein protruding from the membrane and in contact with the cytosol or extracellular fluid.
Peripheral proteins are found on either the exterior or interior surfaces of membranes; and weakly or temporarily associated with the membranes. They can interact with either integral membrane proteins or simply interact weakly with the phospholipids within the membrane.

Figure 5. Integral membranes proteins may have one or more α-helices (pink cylinders) that span the membrane (examples 1 and 2), or they may have β-sheets (blue rectangles) that span the membrane (example 3). (credit: “Foobar”/Wikimedia Commons)
Carbohydrates
Carbohydrates are the third major component of plasma membranes. They are always found on the exterior surface of cells and are bound either to proteins (forming glycoproteins) or to lipids (forming glycolipids). These carbohydrate chains may consist of 2–60 monosaccharide units and can be either straight or branched. Along with peripheral proteins, carbohydrates form specialized sites on the cell surface that allow cells to recognize each other (one of the core functional requirements noted above in "cellular membranes").
Membrane fluidity
The mosaic characteristic of the membrane, described in the fluid mosaic model, helps to illustrate its nature. The integral proteins and lipids exist in the membrane as separate molecules and they "float" in the membrane, moving somewhat with respect to one another. The membrane is not like a balloon, however, in that can expand and contract dramatically; rather, it is fairly rigid and can burst if penetrated or if a cell takes in too much water. However, because of its mosaic nature, a very fine needle can easily penetrate a plasma membrane without causing it to burst, and the membrane will flow and self-seal when the needle is extracted.
The mosaic characteristics of the membrane explain some but not all of its fluidity. There are two other factors that help maintain this fluid characteristic. One factor is the nature of the phospholipids themselves. In their saturated form, the fatty acids in phospholipid tails are saturated with hydrogen atoms. There are no double bonds between adjacent carbon atoms. This results in tails that are relatively straight. By contrast, unsaturated fatty acids do not have a full complement of hydrogen atoms on their fatty acid tails, and therefore contain some double bonds between adjacent carbon atoms; a double bond results in a bend in the string of carbons of approximately 30 degrees.

Figure 6. Any given cell membrane will be composed of a combination of saturated and unsaturated phospholipids. The ratio of the two will influence the permeability and fluidity of the membrane. A membrane composed of completely saturated lipids will be dense and less fluid, and a membrane composed of completely unsaturated lipids will be very loose and very fluid.
Note: possible discussion
Organisms can be found living in extreme temperature conditions. Both in extreme cold or extreme heat. What types of differences would you expect to see in the lipid composition of organisms that live at these extremes?
Saturated fatty acids, with straight tails, are compressed by decreasing temperatures, and they will press in on each other, making a dense and fairly rigid membrane. When unsaturated fatty acids are compressed, the “kinked” tails elbow adjacent phospholipid molecules away, maintaining some space between the phospholipid molecules. This “elbow room” helps to maintain fluidity in the membrane at temperatures at which membranes with high concentrations of saturated fatty acid tails would “freeze” or solidify. The relative fluidity of the membrane is particularly important in a cold environment. Many organisms (fish are one example) are capable of adapting to cold environments by changing the proportion of unsaturated fatty acids in their membranes in response to the lowering of the temperature.
Cholesterol
Animals have an additional membrane constituent that assists in maintaining fluidity. Cholesterol, which lies alongside the phospholipids in the membrane, tends to dampen the effects of temperature on the membrane. Thus, this lipid functions as a "fluidity buffer", preventing lower temperatures from inhibiting fluidity and preventing increased temperatures from increasing fluidity too much. Thus, cholesterol extends, in both directions, the range of temperature in which the membrane is appropriately fluid and consequently functional. Cholesterol also serves other functions, such as organizing clusters of transmembrane proteins into lipid rafts.

Figure 7. Cholesterol fits between the phospholipid groups within the membrane.
Review of the components of the membrane
While there are certain trends or chemical properties that can be roughly associated with different compound permeability (small thing go through "fast", big things "slowly", charged things not at all etc.), we caution against over-generalizing. The molecular determinants of membrane permeability are complicated and involve numerous factors including: the specific composition of the membrane, temperature, ionic composition, hydration; the chemical properties of the solute; the potential chemical interactions between the solute in solution and in the membrane; the dielectric properties of materials; and the energy trade-offs associated with moving substances into and out of various environments. So, in this class, rather than try to apply "rules" and try to develop too many arbitrary "cut-offs", we will strive to develop a general sense of some properties that can influence permeability and leave the assignment of absolute permeability to experimentally reported rates. In addition, we will also try to minimize the use of vocabulary that depends on a frame of reference. For instance, saying that compound A diffuses "quickly" or "slowly" across a bilayer only means something if the terms "quickly" or "slowly" are numerically defined or the biological context is understood.
Energetics of transport
All substances that move through the membrane do so by one of two general methods, which are categorized based on whether or not the transport process is exergonic or endergonic. Passive transport is the exergonic movement of substances across the membrane. In contrast, active transport is the endergonic movement of substances across the membrane that is coupled to an exergonic reaction.
Passive transport
Passive transport does not require the cell to expend energy. In passive transport, substances move from an area of higher concentration to an area of lower concentration, down their concentration gradient . Depending on the chemical nature of the substance, different processes may be associated with passive transport.
Diffusion
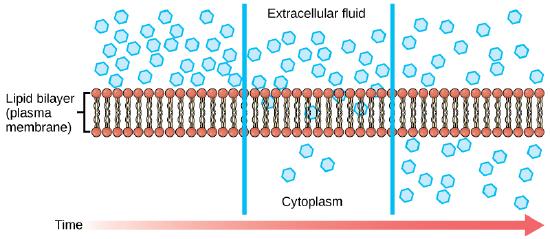
Diffusion is a passive process of transport. A single substance tends to move from an area of high concentration to an area of low concentration until the concentration is equal across a space. You are familiar with diffusion of substances through the air. For example, think about someone opening a bottle of ammonia in a room filled with people. The ammonia gas is at its highest concentration in the bottle; its lowest concentration is at the edges of the room. The ammonia vapor will diffuse, or spread away, from the bottle; gradually, more and more people will smell the ammonia as it spreads. Materials move within the cell’s cytosol by diffusion, and certain materials move through the plasma membrane by diffusion.
Factors that affect diffusion
If unconstrained, molecules will move through and explore space randomly at a rate that depends on their size, their shape, their environment, and their thermal energy. This type of movement underlies the diffusive movement of molecules through whatever medium they are in. The absence of a concentration gradient does not mean that this movement will stop, just that there may be no net movement of the number of molecules from one area to another, a condition known as dynamic equilibrium.
Factors influencing diffusion include:
- Extent of the concentration gradient: The greater the difference in concentration, the more rapid the diffusion. The closer the distribution of the material gets to equilibrium, the slower the rate of diffusion becomes.
- Shape, size and mass of the molecules diffusing: Large and heavier molecules move more slowly; therefore, they diffuse more slowly. The reverse is typically true for smaller, lighter molecules.
- Temperature: Higher temperatures increase the energy and therefore the movement of the molecules, increasing the rate of diffusion. Lower temperatures decrease the energy of the molecules, thus decreasing the rate of diffusion.
- Solvent density: As the density of a solvent increases, the rate of diffusion decreases. The molecules slow down because they have a more difficult time getting through the denser medium. If the medium is less dense, rates of diffusion increase. Since cells primarily use diffusion to move materials within the cytoplasm, any increase in the cytoplasm’s density will decrease the rate at which materials move in the cytoplasm.
- Solubility: As discussed earlier, nonpolar or lipid-soluble materials pass through plasma membranes more easily than polar materials, allowing a faster rate of diffusion.
- Surface area and thickness of the plasma membrane: Increased surface area increases the rate of diffusion, whereas a thicker membrane reduces it.
- Distance traveled: The greater the distance that a substance must travel, the slower the rate of diffusion. This places an upper limitation on cell size. A large, spherical cell will die because nutrients or waste cannot reach or leave the center of the cell, respectively. Therefore, cells must either be small in size, as in the case of many prokaryotes, or be flattened, as with many single-celled eukaryotes.
Facilitated transport
In facilitated transport, also called facilitated diffusion, materials diffuse across the plasma membrane with the help of membrane proteins. A concentration gradient exists that allows these materials to diffuse into or out of the cell without expending cellular energy. In the case that the materials are ions or polar molecules (compounds that are repelled by the hydrophobic parts of the cell membrane), facilitated transport proteins help shield these materials from the repulsive force of the membrane, allowing them to diffuse into the cell.
Note: possible discussion
Compare and contrast passive diffusion and facilitated diffusion.
Channels
The integral proteins involved in facilitated transport are collectively referred to as transport proteins, and they function as either channels for the material or carriers. In both cases, they are transmembrane proteins. Different channel proteins have different transport properties. Some have evolved to have very high specificity for the substance that is being transported while others transport a variety of molecules sharing some common characteristic(s). The interior "passageway" of channel proteins have evolved to provide a low energetic barrier for transport of substances across the membrane through the complementary arrangement of amino acid functional groups (of both backbone and side-chains). Passage through the channel allows polar compounds to avoid the nonpolar central layer of the plasma membrane that would otherwise slow or prevent their entry into the cell. While at any one time significant amounts of water crosses the membrane both in and out, the rate of individual water molecule transport may not be fast enough to adapt to changing environmental conditions. For such cases Nature has evolved a special class of membrane proteins called aquaporins that allow water to pass through the membrane at a very high rate.
Channel proteins are either open at all times or they are “gated.” The latter controls the opening of the channel. Various mechanisms may be involved in the gating mechanism. For instance, the attachment of a specific ion or small molecule to the channel protein may trigger opening. Changes in local membrane "stress" or changes in voltage across the membrane may also be triggers to open or close a channel.
Different organisms and tissues in multicellular species express different sets of channel proteins in their membranes depending on the environments they live in or specialized function they play in an organisms. This provides each type of cell with a unique membrane permeability profile that is evolved to complement its "needs" (note the anthropomorphism). For example, in some tissues, sodium and chloride ions pass freely through open channels, whereas in other tissues a gate must be opened to allow passage. This occurs in the kidney, where both forms of channels are found in different parts of the renal tubules. Cells involved in the transmission of electrical impulses, such as nerve and muscle cells, have gated channels for sodium, potassium, and calcium in their membranes. Opening and closing of these channels changes the relative concentrations on opposing sides of the membrane of these ions, resulting a change in electrical potential across the membrane that lead to message propagation in the case of nerve cells or in muscle contraction in the case of muscle cells.
Carrier proteins
Another type of protein embedded in the plasma membrane is a carrier protein. This aptly named protein binds a substance and, in doing so, triggers a change of its own shape, moving the bound molecule from the outside of the cell to its interior; depending on the gradient, the material may move in the opposite direction. Carrier proteins are typically specific for a single substance. This selectivity adds to the overall selectivity of the plasma membrane. The molecular-scale mechanism of function for these proteins remains poorly understood.
Carrier protein play an important role in the function of kidneys. Glucose, water, salts, ions, and amino acids needed by the body are filtered in one part of the kidney. This filtrate, which includes glucose, is then reabsorbed in another part of the kidney with the help of carrier proteins. Because there are only a finite number of carrier proteins for glucose, if more glucose is present in the filtrate than the proteins can handle, the excess is not reabsorbed and it is excreted from the body in the urine. In a diabetic individual, this is described as “spilling glucose into the urine.” A different group of carrier proteins called glucose transport proteins, or GLUTs, are involved in transporting glucose and other hexose sugars through plasma membranes within the body.
Channel and carrier proteins transport material at different rates. Channel proteins transport much more quickly than do carrier proteins. Channel proteins facilitate diffusion at a rate of tens of millions of molecules per second, whereas carrier proteins work at a rate of a thousand to a million molecules per second.
Active transport
Active transport mechanisms require the use of the cell’s energy, usually in the form of adenosine triphosphate (ATP). If a substance must move into the cell against its concentration gradient—that is, if the concentration of the substance inside the cell is greater than its concentration in the extracellular fluid (and vice versa)—the cell must use energy to move the substance. Some active transport mechanisms move small-molecular weight materials, such as ions, through the membrane. Other mechanisms transport much larger molecules.
Moving against a gradient
To move substances against a concentration or electrochemical gradient, the cell must use energy. This energy is harvested from ATP generated through the cell’s metabolism. Active transport mechanisms, collectively called pumps, work against electrochemical gradients. Small substances constantly pass through plasma membranes. Active transport maintains concentrations of ions and other substances needed by living cells in the face of these passive movements. Much of a cell’s supply of metabolic energy may be spent maintaining these processes. (Most of a red blood cell’s metabolic energy is used to maintain the imbalance between exterior and interior sodium and potassium levels required by the cell.) Because active transport mechanisms depend on a cell’s metabolism for energy, they are sensitive to many metabolic poisons that interfere with the supply of ATP.
Two mechanisms exist for the transport of small-molecular weight material and small molecules. Primary active transport moves ions across a membrane and creates a difference in charge across that membrane, which is directly dependent on ATP. Secondary active transport describes the movement of material that is due to the electrochemical gradient established by primary active transport that does not directly require ATP.
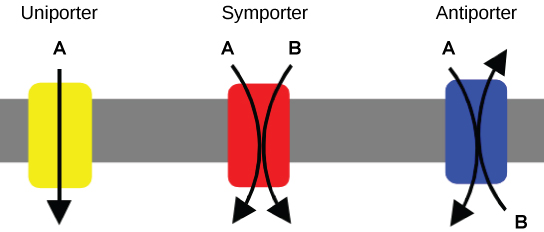
Carrier proteins for active transport
An important membrane adaption for active transport is the presence of specific carrier proteins or pumps to facilitate movement: there are three types of these proteins or transporters. A uniporter carries one specific ion or molecule. A symporter carries two different ions or molecules, both in the same direction. An antiporter also carries two different ions or molecules, but in different directions. All of these transporters can also transport small, uncharged organic molecules like glucose. These three types of carrier proteins are also found in facilitated diffusion, but they do not require ATP to work in that process. Some examples of pumps for active transport are Na+-K+ ATPase, which carries sodium and potassium ions, and H+-K+ ATPase, which carries hydrogen and potassium ions. Both of these are antiporter carrier proteins. Two other carrier proteins are Ca2+ATPase and H+ ATPase, which carry only calcium and only hydrogen ions, respectively. Both are pumps.
Primary active transport
In primary active transport, the energy is often - though not exclusively - derived directly from the hydrolysis of ATP. Often, primary active transport, such as that shown below, which functions to transport sodium and potassium ions allows secondary active transport to occur (discussed in the section below). The second transport method is still considered active because it depends on the use of energy from the primary transport.
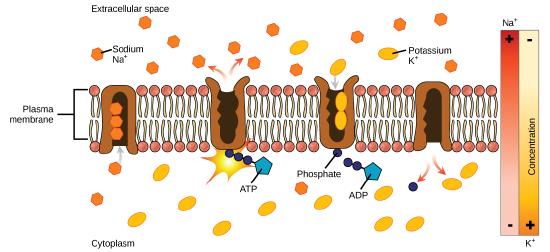
One of the most important pumps in animal cells is the sodium-potassium pump (Na+-K+ ATPase), which maintains the electrochemical gradient (and the correct concentrations of Na+and K+) in living cells. The sodium-potassium pump moves K+ into the cell while moving Na+ out at the same time, at a ratio of three Na+ for every two K+ ions moved in. The Na+-K+ATPase exists in two forms depending on its orientation to the interior or exterior of the cell and its affinity for either sodium or potassium ions. The process consists of the following six steps.
- With the enzyme oriented towards the interior of the cell, the carrier has a high affinity for sodium ions. Three ions bind to the protein.
- ATP is hydrolyzed by the protein carrier and a low-energy phosphate group attaches to it.
- As a result, the carrier changes shape and re-orients itself towards the exterior of the membrane. The protein’s affinity for sodium decreases and the three sodium ions leave the carrier.
- The shape change increases the carrier’s affinity for potassium ions, and two such ions attach to the protein. Subsequently, the low-energy phosphate group detaches from the carrier.
- With the phosphate group removed and potassium ions attached, the carrier protein repositions itself towards the interior of the cell.
- The carrier protein, in its new configuration, has a decreased affinity for potassium, and the two ions are released into the cytoplasm. The protein now has a higher affinity for sodium ions, and the process starts again.
Several things have happened as a result of this process. At this point, there are more sodium ions outside of the cell than inside and more potassium ions inside than out. For every three ions of sodium that move out, two ions of potassium move in. This results in the interior being slightly more negative relative to the exterior. This difference in charge is important in creating the conditions necessary for the secondary process. The sodium-potassium pump is, therefore, an electrogenic pump (a pump that creates a charge imbalance), creating an electrical imbalance across the membrane and contributing to the membrane potential.
Link to learning
Visit the site to see a simulation of active transport in a sodium-potassium ATPase.
Secondary active transport (co-transport)
Secondary active transport brings sodium ions, and possibly other compounds, into the cell. As sodium ion concentrations build outside of the plasma membrane because of the action of the primary active transport process, an electrochemical gradient is created. If a channel protein exists and is open, the sodium ions will be pulled through the membrane. This movement is used to transport other substances that can attach themselves to the transport protein through the membrane. Many amino acids, as well as glucose, enter a cell this way. This secondary process is also used to store high energy hydrogen ions in the mitochondria of plant and animal cells for the production of ATP. The potential energy that accumulates in the stored hydrogen ions is translated into kinetic energy as the ions surge through the channel protein ATP synthase, and that energy is used to convert ADP into ATP.

Osmosis
Osmosis is the movement of water through a semipermeable membrane according to the concentration gradient of water across the membrane, which is inversely proportional to the concentration of solutes. While diffusion transports material across membranes and within cells, osmosis transports only water across a membrane and the membrane limits the diffusion of solutes in the water. Not surprisingly, the aquaporins that facilitate water movement play a large role in osmosis, most prominently in red blood cells and the membranes of kidney tubules.
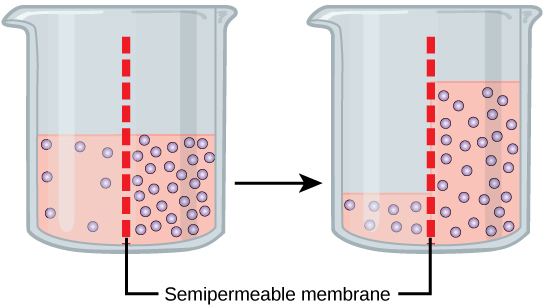
Mechanism
Osmosis is a special case of diffusion. Water, like other substances, moves from an area of high concentration to one of low concentration. An obvious question is what makes water move at all? Imagine a beaker with a semipermeable membrane separating the two sides or halves. On both sides of the membrane the water level is the same, but there are different concentrations of a dissolved substance, or solute, that cannot cross the membrane (otherwise the concentrations on each side would be balanced by the solute crossing the membrane). If the volume of the solution on both sides of the membrane is the same, but the concentrations of solute are different, then there are different amounts of water, the solvent, on either side of the membrane.
To illustrate this, imagine two full glasses of water. One has a single teaspoon of sugar in it, whereas the second one contains one-quarter cup of sugar. If the total volume of the solutions in both cups is the same, which cup contains more water? Because the large amount of sugar in the second cup takes up much more space than the teaspoon of sugar in the first cup, the first cup has more water in it.
Returning to the beaker example, recall that it has a mixture of solutes on either side of the membrane. A principle of diffusion is that the molecules move around and will spread evenly throughout the medium if they can. However, only the material capable of getting through the membrane will diffuse through it. In this example, the solute cannot diffuse through the membrane, but the water can. Water has a concentration gradient in this system. Thus, water will diffuse down its concentration gradient, crossing the membrane to the side where it is less concentrated. This diffusion of water through the membrane—osmosis—will continue until the concentration gradient of water goes to zero or until the hydrostatic pressure of the water balances the osmotic pressure. Osmosis proceeds constantly in living systems.
Tonicity
Tonicity describes how an extracellular solution can change the volume of a cell by affecting osmosis. A solution's tonicity often directly correlates with the osmolarity of the solution. Osmolarity describes the total solute concentration of the solution. A solution with low osmolarity has a greater number of water molecules relative to the number of solute particles; a solution with high osmolarity has fewer water molecules with respect to solute particles. In a situation in which solutions of two different osmolarities are separated by a membrane permeable to water, though not to the solute, water will move from the side of the membrane with lower osmolarity (and more water) to the side with higher osmolarity (and less water). This effect makes sense if you remember that the solute cannot move across the membrane, and thus the only component in the system that can move—the water—moves along its own concentration gradient. An important distinction that concerns living systems is that osmolarity measures the number of particles (which may be molecules) in a solution. Therefore, a solution that is cloudy with cells may have a lower osmolarity than a solution that is clear if the second solution contains more dissolved molecules than there are cells.
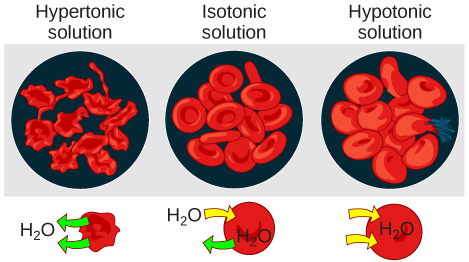
Hypotonic solutions
Three terms—hypotonic, isotonic, and hypertonic—are used to relate the osmolarity of a cell to the osmolarity of the extracellular fluid that contains the cells. In a hypotonic situation, the extracellular fluid has lower osmolarity than the fluid inside the cell, and water enters the cell (in living systems, the point of reference is always the cytoplasm, so the prefix hypo- means that the extracellular fluid has a lower concentration of solutes, or a lower osmolarity, than the cell cytoplasm). It also means that the extracellular fluid has a higher concentration of water in the solution than does the cell. In this situation, water will follow its concentration gradient and enter the cell.
Hypertonic solutions
As for a hypertonic solution, the prefix hyper- refers to the extracellular fluid having a higher osmolarity than the cell’s cytoplasm; therefore, the fluid contains less water than the cell does. Because the cell has a relatively higher concentration of water, water will leave the cell.
Isotonic solutions
In an isotonic solution, the extracellular fluid has the same osmolarity as the cell. If the osmolarity of the cell matches that of the extracellular fluid, there will be no net movement of water into or out of the cell, although water will still move in and out. Blood cells and plant cells in hypertonic, isotonic, and hypotonic solutions take on characteristic appearances.
Connection
A doctor injects a patient with what the doctor thinks is an isotonic saline solution. The patient dies, and an autopsy reveals that many red blood cells have been destroyed. Do you think the solution the doctor injected was really isotonic?
Link to learning
For a video illustrating the process of diffusion in solutions, visit this site.
Tonicity in living systems
In a hypotonic environment, water enters a cell, and the cell swells. In an isotonic condition, the relative concentrations of solute and solvent are equal on both sides of the membrane. There is no net water movement; therefore, there is no change in the size of the cell. In a hypertonic solution, water leaves a cell and the cell shrinks. If either the hypo- or hyper- condition goes to excess, the cell’s functions become compromised, and the cell may be destroyed.
A red blood cell will burst, or lyse, when it swells beyond the plasma membrane’s capability to expand. Remember, the membrane resembles a mosaic, with discrete spaces between the molecules composing it. If the cell swells, and the spaces between the lipids and proteins become too large, and the cell will break apart.
In contrast, when excessive amounts of water leave a red blood cell, the cell shrinks, or crenates. This has the effect of concentrating the solutes left in the cell, making the cytosol denser and interfering with diffusion within the cell. The cell’s ability to function will be compromised and may also result in the death of the cell.
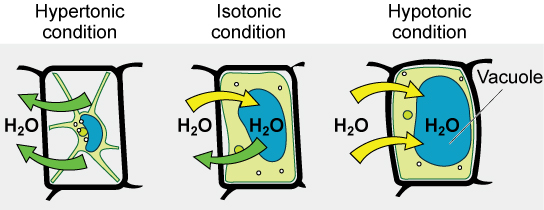
Various living things have ways of controlling the effects of osmosis—a mechanism called osmoregulation. Some organisms, such as plants, fungi, bacteria, and some protists, have cell walls that surround the plasma membrane and prevent cell lysis in a hypotonic solution. The plasma membrane can only expand to the limit of the cell wall, so the cell will not lyse. In fact, the cytoplasm in plants is always slightly hypertonic to the cellular environment, and water will always enter a cell if water is available. This inflow of water produces turgor pressure, which stiffens the cell walls of the plant. In nonwoody plants, turgor pressure supports the plant. Conversely, if the plant is not watered, the extracellular fluid will become hypertonic, causing water to leave the cell. In this condition, the cell does not shrink because the cell wall is not flexible. However, the cell membrane detaches from the wall and constricts the cytoplasm. This is called plasmolysis. Plants lose turgor pressure in this condition and wilt.
Tonicity is a concern for all living things. For example, paramecia and amoebas, which are protists that lack cell walls, have contractile vacuoles. This vesicle collects excess water from the cell and pumps it out, keeping the cell from bursting as it takes on water from its environment.
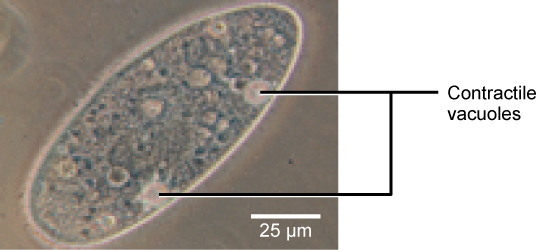
Figure 12. A paramecium’s contractile vacuole, here visualized using bright field light microscopy at 480x magnification, continuously pumps water out of the organism’s body to keep it from bursting in a hypotonic medium. (credit: modification of work by NIH; scale-bar data from Matt Russell)
Many marine invertebrates have internal salt levels matched to their environments, making them isotonic with the water in which they live. Fish, however, must spend approximately five percent of their metabolic energy maintaining osmotic homeostasis. Freshwater fish live in an environment that is hypotonic to their cells. These fish actively take in salt through their gills and excrete diluted urine to rid themselves of excess water. Saltwater fish live in the reverse environment, which is hypertonic to their cells, and they secrete salt through their gills and excrete highly concentrated urine.
In vertebrates, the kidneys regulate the amount of water in the body. Osmoreceptors are specialized cells in the brain that monitor the concentration of solutes in the blood. If the levels of solutes increase beyond a certain range, a hormone is released that retards water loss through the kidney and dilutes the blood to safer levels. Animals also have high concentrations of albumin, which is produced by the liver, in their blood. This protein is too large to pass easily through plasma membranes and is a major factor in controlling the osmotic pressures applied to tissues.
Eukaryotes
Living things fall into three large groups: Archaea, Bacteria, and Eukarya. The first two groups include non-nucleated cells, and the third contains all eukaryotes. A relatively sparse fossil record is available to help us discern what the first members of each of these lineages looked like, so it is possible that all the events that led up to the last common ancestor of extant eukaryotes will remain unknown. However, comparative biology of extant organisms and the limited fossil record provide some insights into the history of Eukarya.
The earliest fossils found appear to be bacteria, most likely cyanobacteria. They are about 3.5 billion years old and are recognizable because of their relatively complex structure and, for bacteria, relatively large cells. Most other bacteria and archaea have small cells, 1 or 2 µm in size, and would be difficult to pick out as fossils. Most living eukaryotes have cells measuring 10 µm or greater. Structures this size, which might be fossils, appear in the geological record about 2.1 billion years ago.
Characteristics of eukaryotes
Data from these fossils have led biologists to the conclusion that living eukaryotes are all descendants of a single common ancestor. Mapping the characteristics found in all major groups of eukaryotes reveals that the following characteristics must have been present in the last common ancestor, because these characteristics are present in at least some of the members of each major lineage.
- Cells with nuclei surrounded by a nuclear envelope with nuclear pores. This is the single characteristic that is both necessary and sufficient to define an organism as a eukaryote. All extant eukaryotes have cells with nuclei.
- Mitochondria. Some extant eukaryotes have very reduced remnants of mitochondria in their cells, whereas other members of their lineages have “typical” mitochondria.
- A cytoskeleton containing the structural and motility components called actin microfilaments and microtubules. All extant eukaryotes have these cytoskeletal elements.
- Flagella and cilia, organelles associated with cell motility. Some extant eukaryotes lack flagella and/or cilia, but they are descended from ancestors that possessed them.
- Chromosomes, each consisting of a linear DNA molecule coiled around basic (alkaline) proteins called histones. The few eukaryotes with chromosomes lacking histones clearly evolved from ancestors that had them.
- Mitosis, a process of nuclear division wherein replicated chromosomes are divided and separated using elements of the cytoskeleton. Mitosis is universally present in eukaryotes.
- Sex, a process of genetic recombination unique to eukaryotes in which diploid nuclei at one stage of the life cycle undergo meiosis to yield haploid nuclei and subsequent karyogamy, a stage where two haploid nuclei fuse together to create a diploid zygote nucleus.
- Members of all major lineages have cell walls, and it might be reasonable to conclude that the last common ancestor could make cell walls during some stage of its life cycle. However, not enough is known about eukaryotes’ cell walls and their development to know how much homology exists among them. If the last common ancestor could make cell walls, it is clear that this ability must have been lost in many groups.
Endosymbiosis and the evolution of eukaryotes
In order to understand eukaryotic organisms fully, it is necessary to understand that all extant eukaryotes are descendants of a chimeric organism that was a composite of a host cell and the cell(s) of an alpha-proteobacterium that “took up residence” inside it. This major theme in the origin of eukaryotes is known as endosymbiosis, one cell engulfing another such that the engulfed cell survives and both cells benefit. Over many generations, a symbiotic relationship can result in two organisms that depend on each other so completely that neither could survive on its own. Endosymbiotic events likely contributed to the origin of the last common ancestor of today’s eukaryotes and to later diversification in certain lineages of eukaryotes. Before explaining this further, it is necessary to consider metabolism in bacteria and archaea.
Bacterial and archaeal metabolism
Many important metabolic processes arose in bacteria and archaea, and some of these, such as nitrogen fixation, are never found in eukaryotes. The process of aerobic respiration is found in all major lineages of eukaryotes, and it is localized in the mitochondria. Aerobic respiration is also found in many lineages of bacteria and archaea, but it is not present in all of them, and many forms of evidence suggest that such anaerobic microbes never carried out aerobic respiration nor did their ancestors.
While today’s atmosphere is about one-fifth molecular oxygen (O2), geological evidence shows that it originally lacked O2. Without oxygen, aerobic respiration would not be expected, and living things would have relied on fermentation instead. At some point around 3.5 billion years ago, some bacteria and archaea began using energy from sunlight to power anabolic processes that reduce carbon dioxide to form organic compounds. That is, they evolved the ability to photosynthesize. Hydrogen, derived from various sources, was captured using light-powered reactions to reduce fixed carbon dioxide in the Calvin cycle. The group of Gram-negative bacteria that gave rise to cyanobacteria used water as the hydrogen source and released O2 as a waste product.
Eventually, the amount of photosynthetic oxygen built up in some environments to levels that posed a risk to living organisms, since it can damage many organic compounds. Various metabolic processes evolved that protected organisms from oxygen; one of which, aerobic respiration, also generated high levels of ATP. It became widely present among microbes, including in a group we now call alpha-proteobacteria. Organisms that did not acquire aerobic respiration had to remain in oxygen-free environments. Originally, oxygen-rich environments were likely localized around places where cyanobacteria were active, but by about 2 billion years ago, geological evidence shows that oxygen was building up to higher concentrations in the atmosphere. Oxygen levels similar to today’s levels only arose within the last 700 million years.
Recall that the first fossils that we believe to be eukaryotes are about 2 billion years old, so they appeared as oxygen levels were increasing. Also, recall that all extant eukaryotes descended from an ancestor with mitochondria. These organelles were first observed by light microscopists in the late 1800s, where they appeared to be somewhat worm-shaped structures that seemed to be moving around in the cell. Some early observers suggested that they might be bacteria living inside host cells, but these hypotheses remained unknown or rejected in most scientific communities.
Endosymbiotic theory
As cell biology developed in the twentieth century, it became clear that mitochondria were the organelles responsible for producing ATP using aerobic respiration. In the 1960s, American biologist Lynn Margulis developed endosymbiotic theory, which states that eukaryotes may have been a product of one cell engulfing another (one living within another) and evolving over time until the separate cells were no longer recognizable as such. In 1967, Margulis introduced new work on the theory and substantiated her findings through microbiological evidence. Although Margulis’ work initially was met with resistance, this once-revolutionary hypothesis is now widely (but not completely) accepted, with work progressing on uncovering the steps involved in this evolutionary process and the key players involved. Much still remains to be discovered about the origins of the cells that now make up the cells in all living eukaryotes.
Broadly, it has become clear that many of our nuclear genes and the molecular machinery responsible for replication and expression appear closely related to those in Archaea. On the other hand, the metabolic organelles and genes responsible for many energy-harvesting processes had their origins in bacteria. Much remains to be clarified about how this relationship occurred; this continues to be an exciting field of discovery in biology. For instance, it is not known whether the endosymbiotic event that led to mitochondria occurred before or after the host cell had a nucleus. Such organisms would be among the extinct precursors of the last common ancestor of eukaryotes.
Mitochondria
One of the major features distinguishing bacteria and archaea from eukaryotes is the presence of mitochondria. Eukaryotic cells may contain anywhere from one to several thousand mitochondria, depending on the cell’s level of energy consumption. Each mitochondrion measures 1 to 10 or greater micrometers in length and exists in the cell as an organelle that can be ovoid, worm-shaped, or intricately branched. Mitochondria arise from the division of existing mitochondria; they may fuse together; and they may be moved around inside the cell by interactions with the cytoskeleton. However, mitochondria cannot survive outside the cell. As the atmosphere was oxygenated by photosynthesis, and as successful aerobic microbes evolved, evidence suggests that an ancestral cell with some membrane compartmentalization engulfed a free-living aerobic bacterium, specifically an alpha-proteobacterium, thereby giving the host cell the ability to use oxygen to release energy stored in nutrients. Alpha-proteobacteria are a large group of bacteria that includes species symbiotic with plants, disease organisms that can infect humans via ticks, and many free-living species that use light for energy. Several lines of evidence support that mitochondria are derived from this endosymbiotic event. Most mitochondria are shaped like alpha-proteobacteria and are surrounded by two membranes, which would result when one membrane-bound organism is engulfed into a vacuole by another membrane-bound organism. The mitochondrial inner membrane is extensive and involves substantial infoldings called cristae that resemble the textured, outer surface of alpha-proteobacteria. The matrix and inner membrane are rich with the enzymes necessary for aerobic respiration.
Eukaryotic Cell: Structure and Function
Introduction to eukaryotic cells
By definition, eukaryotic cells are cells that contain a membrane-bound nucleus, a structural feature that is not present in bacterial or archaeal cells. In addition to the nucleus, eukaryotic cells are characterized by numerous membrane-bound organelles such as the endoplasmic reticulum, Golgi apparatus, chloroplasts, mitochondria, and others.
In previous sections, we began to consider the Design Challenge of making cells larger than a small bacterium—more precisely, growing cells to sizes at which, in the eyes of natural selection, relying on diffusion of substances for transport through a highly viscous cytosol comes with inherent functional trade-offs that offset most selective benefits of getting larger. In the lectures and readings on bacterial cell structure, we discovered some morphological features of large bacteria that allow them to effectively overcome diffusion-limited size barriers (e.g., filling the cytoplasm with a large storage vacuole maintains a small volume for metabolic activity that remains compatible with diffusion-driven transport).
As we transition our focus to eukaryotic cells, we want you to approach the study by constantly returning to the Design Challenge. We will cover a large number of subcellular structures that are unique to eukaryotes, and you will certainly be expected to know the names of these structures or organelles, to associate them with one or more "functions", and to identify them on a canonical cartoon representation of a eukaryotic cell. This memorization exercise is necessary but not sufficient. We will also ask you to start thinking a bit deeper about some of the functional and evolutionary costs and benefits (trade-offs) of both evolving eukaryotic cells and various eukaryotic organelles, as well as how a eukaryotic cell might coordinate the functions of different organelles.
Your instructors will, of course, propose some functional hypotheses for you to consider that address these broader points. Our hypotheses may sometimes come in the form of statements like, "Thing A exists because of rationale B." To be completely honest, however, in many cases, we don't actually know all of the selective pressures that led to the creation or maintenance of certain cellular structures, and the likelihood that one explanation will fit all cases is slim in biology. The causal linkage/relationship implied by the use of terms like "because" should be treated as good hypotheses rather than objective, concrete, undisputed, factual knowledge. We want you to understand these hypotheses and to be able to discuss the ideas presented in class, but we also want you to indulge your own curiosity and to begin thinking critically about these ideas yourself. Try using the Design Challenge rubric to explore some of your ideas. In the following, we will try to seed questions to encourage this activity.

Figure 1. These figures show the major organelles and other cell components of (a) a typical animal cell and (b) a typical eukaryotic plant cell. The plant cell has a cell wall, chloroplasts, plastids, and a central vacuole—structures not found in animal cells. Plant cells do not have lysosomes or centrosomes.
The plasma membrane
Like bacteria and archaea, eukaryotic cells have a plasma membrane, a phospholipid bilayer with embedded proteins that separates the internal contents of the cell from its surrounding environment. The plasma membrane controls the passage of organic molecules, ions, water, and oxygen into and out of the cell. Wastes (such as carbon dioxide and ammonia) also leave the cell by passing through the plasma membrane, usually with some help of protein transporters.

Figure 2. The eukaryotic plasma membrane is a phospholipid bilayer with proteins and cholesterol embedded in it.
As discussed in the context of bacterial cell membranes, the plasma membranes of eukaryotic cells may also adopt unique conformations. For instance, the plasma membrane of cells that, in multicellular organisms, specialize in absorption are often folded into fingerlike projections called microvilli (singular = microvillus); (see figure below). The "folding" of the membrane into microvilli effectively increases the surface area for absorption while minimally impacting the cytosolic volume. Such cells can be found lining the small intestine, the organ that absorbs nutrients from digested food.
An aside: People with celiac disease have an immune response to gluten, a protein found in wheat, barley, and rye. The immune response damages microvilli. As a consequence, afflicted individuals have an impaired ability to absorb nutrients. This can lead to malnutrition, cramping, and diarrhea.

Figure 3. Microvilli, shown here as they appear on cells lining the small intestine, increase the surface area available for absorption. These microvilli are only found on the area of the plasma membrane that faces the cavity from which substances will be absorbed. Credit: "micrograph", modification of work by Louisa Howard
The cytoplasm
The cytoplasm refers to the entire region of a cell between the plasma membrane and the nuclear envelope. It is composed of organelles suspended in the gel-like cytosol, the cytoskeleton, and various chemicals (see figure below). Even though the cytoplasm consists of 70 to 80 percent water, it nevertheless has a semisolid consistency. It is crowded in there. Proteins, simple sugars, polysaccharides, amino acids, nucleic acids, fatty acids, ions and many other water-soluble molecules are all competing for space and water.
The nucleus
Typically, the nucleus is the most prominent organelle in a cell (see figure below) when viewed through a microscope. The nucleus (plural = nuclei) houses the cell’s DNA. Let’s look at it in more detail.

Figure 4. The nucleus stores chromatin (DNA plus proteins) in a gel-like substance called the nucleoplasm. The nucleolus is a condensed region of chromatin where ribosome synthesis occurs. The boundary of the nucleus is called the nuclear envelope. It consists of two phospholipid bilayers: an outer membrane and an inner membrane. The nuclear membrane is continuous with the endoplasmic reticulum. Nuclear pores allow substances to enter and exit the nucleus.
The nuclear envelope
The nuclear envelope, a structure that constitutes the outermost boundary of the nucleus, is a double-membrane—both the inner and outer membranes of the nuclear envelope are phospholipid bilayers. The nuclear envelope is also punctuated with protein-based pores that control the passage of ions, molecules, and RNA between the nucleoplasm and cytoplasm. The nucleoplasm is the semisolid fluid inside the nucleus where we find the chromatin and the nucleolus, a condensed region of chromatin where ribosome synthesis occurs.
Chromatin and chromosomes
To understand chromatin, it is helpful to first consider chromosomes. Chromosomes are structures within the nucleus that are made up of DNA, the hereditary material. You may remember that in bacteria and archaea, DNA is typically organized into one or more circular chromosome(s). In eukaryotes, chromosomes are linear structures. Every eukaryotic species has a specific number of chromosomes in the nuclei of its cells. In humans, for example, the chromosome number is 23, while in fruit flies, it is 4.
Chromosomes are only clearly visible and distinguishable from one another by visible optical microscopy when the cell is preparing to divide and the DNA is tightly packed by proteins into easily distinguishable shapes. When the cell is in the growth and maintenance phases of its life cycle, numerous proteins are still associated with the nucleic acids, but the DNA strands more closely resemble an unwound, jumbled bunch of threads. The term chromatin is used to describe chromosomes (the protein-DNA complexes) when they are both condensed and decondensed.

Figure 5. (a) This image shows various levels of the organization of chromatin (DNA and protein). (b) This image shows paired chromosomes. Credit (b): modification of work by NIH; scale-bar data from Matt Russell
The nucleolus
Some chromosomes have sections of DNA that encode ribosomal RNA. A darkly staining area within the nucleus called the nucleolus (plural = nucleoli) aggregates the ribosomal RNA with associated proteins to assemble the ribosomal subunits that are then transported out to the cytoplasm through the pores in the nuclear envelope.
Note: possible discussion
Discuss amongst yourselves. Use the Design Challenge rubric to consider the nucleus in more detail. What "problems" does an organelle like the nucleus solve? What are some of the qualities of a nucleus that may be responsible for ensuring its evolutionary success? What are some of the trade-offs of evolving and maintaining a nucleus? (Every benefit has some cost; can you list both?) Remember, there may be some well-established hypotheses (and it is good to mention these), but the point of the exercise here is for you to think critically and to critically discuss these ideas using your collective "smarts".
Ribosomes
Ribosomes are the cellular structures responsible for protein synthesis. When viewed through an electron microscope, ribosomes appear either as clusters (polyribosomes) or single, tiny dots that float freely in the cytoplasm. They may be attached to the cytoplasmic side of the plasma membrane or the cytoplasmic side of the endoplasmic reticulum and the outer membrane of the nuclear envelope (cartoon of cell above).
Electron microscopy has shown us that ribosomes, which are large complexes of protein and RNA, consist of two subunits, aptly called large and small (figure below). Ribosomes receive their "instructions" for protein synthesis from the nucleus, where the DNA is transcribed into messenger RNA (mRNA). The mRNA travels to the ribosomes, which translate the code provided by the sequence of the nitrogenous bases in the mRNA into a specific order of amino acids in a protein. This is covered in greater detail in the section covering the process of translation.

Figure 6. Ribosomes are made up of a large subunit (top) and a small subunit (bottom). During protein synthesis, ribosomes assemble amino acids into proteins.
Mitochondria
Mitochondria (singular = mitochondrion) are often called the “powerhouses” or “energy factories” of a cell because they are the primary site of metabolic respiration in eukaryotes. Depending on the species and the type of mitochondria found in those cells, the respiratory pathways may be anaerobic or aerobic. By definition, when respiration is aerobic, the terminal electron is oxygen; when respiration is anaerobic, a compound other than oxygen functions as the terminal electron acceptor. In either case, the result of these respiratory processes is the production of ATP via oxidative phosphorylation, hence the use of terms "powerhouse" and/or "energy factory" to describe this organelle. Nearly all mitochondria also possess a small genome that encodes genes whose functions are typically restricted to the mitochondrion.
In some cases, the number of mitochondria per cell is tunable, depending, typically, on energy demand. It is for instance possible muscle cells that are used—that by extension have a higher demand for ATP—may often be found to have a significantly higher number of mitochondria than cells that do not have a high energy load.
The structure of the mitochondria can vary significantly depending on the organism and the state of the cell cycle which one is observing. The typical textbook image, however, depicts mitochondria as oval-shaped organelles with a double inner and outer membrane (see figure below); learn to recognize this generic representation. Both the inner and outer membranes are phospholipid bilayers embedded with proteins that mediate transport across them and catalyze various other biochemical reactions. The inner membrane layer has folds called cristae that increase the surface area into which respiratory chain proteins can be embedded. The region within the cristae is called the mitochondrial matrix and contains—among other things—enzymes of the TCA cycle. During respiration, protons are pumped by respiratory chain complexes from the matrix into a region known as the intermembrane space (between the inner and outer membranes).

Figure 7. This electron micrograph shows a mitochondrion as viewed with a transmission electron microscope. This organelle has an outer membrane and an inner membrane. The inner membrane contains folds, called cristae, which increase its surface area. The space between the two membranes is called the intermembrane space, and the space inside the inner membrane is called the mitochondrial matrix. ATP synthesis takes place on the inner membrane. Credit: modification of work by Matthew Britton; scale-bar data from Matt Russell
Note: possible discussion
Discuss: Processes like glycolysis, lipid biosynthesis, and nucleotide biosynthesis all have compounds that feed into the TCA cycle—some of which occurs in the mitochondria. What are some of the functional challenges associated with coordinating processes that have a common set of molecules if the enzymes are sequestered into different cellular compartments?
Peroxisomes
Peroxisomes are small, round organelles enclosed by single membranes. These organelles carry out redox reactions that oxidize and break down fatty acids and amino acids. They also help to detoxify many toxins that may enter the body. Many of these redox reactions release hydrogen peroxide, H2O2, which would be damaging to cells; however, when these reactions are confined to peroxisomes, enzymes safely break down the H2O2 into oxygen and water. For example, alcohol is detoxified by peroxisomes in liver cells. Glyoxysomes, which are specialized peroxisomes in plants, are responsible for converting stored fats into sugars.
Vesicles and vacuoles
Vesicles and vacuoles are membrane-bound sacs that function in storage and transport. Other than the fact that vacuoles are somewhat larger than vesicles, there is a very subtle distinction between them: the membranes of vesicles can fuse with either the plasma membrane or other membrane systems within the cell. Additionally, some agents such as enzymes within plant vacuoles break down macromolecules. The membrane of a vacuole does not fuse with the membranes of other cellular components.
Animal cells versus plant cells
At this point, you know that each eukaryotic cell has a plasma membrane, cytoplasm, a nucleus, ribosomes, mitochondria, peroxisomes, and in some, vacuoles. There are some striking differences between animal and plant cells worth noting. Here is a brief list of differences that we want you to be familiar with and a slightly expanded description below:
1. While all eukaryotic cells use microtubule and motor protein the based mechanisms to segregate chromosomes during cell division, the structures used to organize these microtubules differ in plants versus animal and yeast cells. Animal and yeast cells organize and anchor their microtubules into structures called microtubule organizing centers (MTOCs). These structures are composed of structures called centrioles that are composed largely of α-tubulin, β-tubulin, and other proteins. Two centrioles organize into a structure called a centrosome. By contrast, in plants, while microtubules also organize into discrete bundles, there are no conspicuous structures similar to the MTOCs seen in animal and yeast cells. Rather, depending on the organism, it appears that there can be several places where these bundles of microtubules can nucleate from places called acentriolar (without centriole) microtubule organizing centers. A third type of tubulin, γ-tubulin, appears to be implicated, but our knowledge of the precise mechanisms used by plants to organize microtubule spindles is still spotty.
2. Animal cells typically have organelles called lysosomes responsible for degradation of biomolecules. Some plant cells contain functionally similar degradative organelles, but there is a debate as to how they should be named. Some plant biologists call these organelles lysosomes while others lump them into the general category of plastids and do not give them a specific name.
3. Plant cells have a cell wall, chloroplasts and other specialized plastids, and a large central vacuole, whereas animal cells do not.
The centrosome
The centrosome is a microtubule-organizing center found near the nuclei of animal cells. It contains a pair of centrioles, two structures that lie perpendicular to eachother (see figure below). Each centriole is a cylinder of nine triplets of microtubules.

Figure 8. The centrosome consists of two centrioles that lie at right angles to each other. Each centriole is a cylinder made up of nine triplets of microtubules. Nontubulin proteins (indicated by the green lines) hold the microtubule triplets together.
The centrosome (the organelle where all microtubules originate in animal and yeast) replicates itself before a cell divides, and the centrioles appear to have some role in pulling the duplicated chromosomes to opposite ends of the dividing cell. However, the exact function of the centrioles in cell division remains unclear, as cells that have had their centrosome removed can still divide, and plant cells, which lack centrosomes, are capable of cell division.
Lysosomes
Animal cells have another set of organelles not found in plant cells: lysosomes. Colloquially, the lysosomes are sometimes called the cell’s “garbage disposal”. Enzymes within the lysosomes aid the breakdown of proteins, polysaccharides, lipids, nucleic acids, and even "worn-out" organelles. These enzymes are active at a much lower pH than that of the cytoplasm. Therefore, the pH within lysosomes is more acidic than the pH of the cytoplasm. In plant cells, many of the same digestive processes take place in vacuoles.
The cell wall
If you examine the diagram above depicting plant and animal cells, you will see in the diagram of a plant cell a structure external to the plasma membrane called the cell wall. The cell wall is a rigid covering that protects the cell, provides structural support, and gives shape to the cell. Fungal and protistan cells also have cell walls. While the chief component of bacterial cell walls is peptidoglycan, the major organic molecule in the plant cell wall is cellulose (see structure below), a polysaccharide made up of glucose subunits.

Figure 9. Cellulose is a long chain of β-glucose molecules connected by a 1-4 linkage. The dashed lines at each end of the figure indicate a series of many more glucose units. The size of the page makes it impossible to portray an entire cellulose molecule.
Chloroplasts
Chloroplasts are plant cell organelles that carry out photosynthesis. Like the mitochondria, chloroplasts have their own DNA and ribosomes, but chloroplasts have an entirely different function.
Like mitochondria, chloroplasts have outer and inner membranes, but within the space enclosed by a chloroplast’s inner membrane is a set of interconnected and stacked fluid-filled membrane sacs called thylakoids (figure below). Each stack of thylakoids is called a granum (plural = grana). The fluid enclosed by the inner membrane that surrounds the grana is called the stroma.

Figure 10. The chloroplast has an outer membrane, an inner membrane, and membrane structures called thylakoids that are stacked into grana. The space inside the thylakoid membranes is called the thylakoid space. The light harvesting reactions take place in the thylakoid membranes, and the synthesis of sugar takes place in the fluid inside the inner membrane, which is called the stroma. Chloroplasts also have their own genome, which is contained on a single circular chromosome.
The chloroplasts contain a green pigment called chlorophyll, which captures the light energy that drives the reactions of photosynthesis. Like plant cells, photosynthetic protists also have chloroplasts. Some bacteria perform photosynthesis, but their chlorophyll is not relegated to an organelle.
Evolution connection
Endosymbiosis
We have mentioned that both mitochondria and chloroplasts contain DNA and ribosomes. Have you wondered why? Strong evidence points to endosymbiosis as the explanation.
Symbiosis is a relationship in which organisms from two separate species depend on each other for their survival. Endosymbiosis (endo- = “within”) is a mutually beneficial relationship in which one organism lives inside the other. Endosymbiotic relationships abound in nature. For instance, some microbes that live in our digestive tracks produce vitamin K. The relationship between these microbes and us (their hosts) is said to be mutually beneficial or symbiotic. The relationship is beneficial for us because we are unable to synthesize vitamin K; the microbes do it for us instead. The relationship is also beneficial for the microbes because they receive abundant food from the environment of the large intestine, and they are protected both from other organisms and from drying out.
Scientists have long noticed that bacteria, mitochondria, and chloroplasts are similar in size. We also know that bacteria have DNA and ribosomes, just as mitochondria and chloroplasts do. Scientists believe that host cells and bacteria formed an endosymbiotic relationship when the host cells ingested both aerobic and autotrophic bacteria (cyanobacteria) but did not destroy them. Through many millions of years of evolution, these ingested bacteria became more specialized in their functions, with the aerobic bacteria becoming mitochondria and the autotrophic bacteria becoming chloroplasts. There will be more on this later in the reading.
The central vacuole
Previously, we mentioned vacuoles as essential components of plant cells. If you look at the cartoon figure of the plant cell, you will see that it depicts a large central vacuole that occupies most of the area of the cell. The central vacuole plays a key role in regulating the cell’s concentration of water in changing environmental conditions.
Silly vacuole factoid: Have you ever noticed that if you forget to water a plant for a few days, it wilts? That’s because as the water concentration in the soil becomes lower than the water concentration in the plant, water moves out of the central vacuoles and cytoplasm. As the central vacuole shrinks, it leaves the cell wall unsupported. This loss of support to the cell walls of plant cells results in the wilted appearance of the plant.
The central vacuole also supports the expansion of the cell. When the central vacuole holds more water, the cell gets larger without having to invest a lot of energy in synthesizing new cytoplasm.
The Endomembrane System
The endomembrane system (endo = “within”) is a group of membranes and organelles in eukaryotic cells that works together to modify, package, and transport lipids and proteins. It includes the nuclear envelope, lysosomes, and vesicles, which we’ve already mentioned, and the endoplasmic reticulum and Golgi apparatus, which we will cover shortly. Although not technically within the cell, the plasma membrane is included in the endomembrane system because, as you will see, it interacts with the other endomembranous organelles. The endomembrane system does not include the membranes of either mitochondria or chloroplasts.
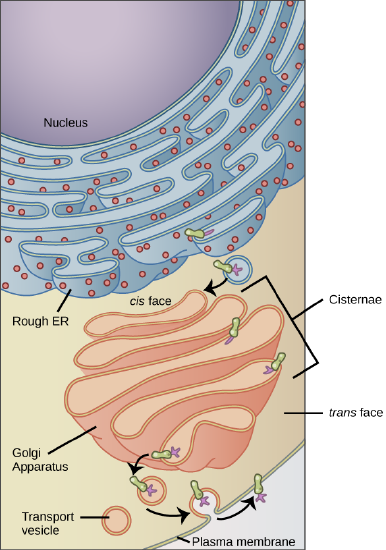
Membrane and secretory proteins are synthesized in the rough endoplasmic reticulum (RER). The RER also sometimes modifies proteins. In this illustration, a (green) integral membrane protein in the ER is modified by attachment of a (purple) carbohydrate. Vesicles with the integral protein bud from the ER and fuse with the cis face of the Golgi apparatus. As the protein passes along the Golgi’s cisternae, it is further modified by the addition of more carbohydrates. After its synthesis is complete, it exits as integral membrane protein of the vesicle that bud from the Golgi’s trans face and when the vesicle fuses with the cell membrane the protein becomes integral portion of that cell membrane. (credit: modification of work by Magnus Manske)
Possible discussion
If a peripheral membrane protein were synthesized in the lumen (inside) of the ER, would it end up on the inside or outside of the plasma membrane?
The Endoplasmic Reticulum
The endoplasmic reticulum (ER) (see figure above) is a series of interconnected membranous sacs and tubules that collectively modifies proteins and synthesizes lipids. However, these two functions are performed in separate areas of the ER: the rough ER and the smooth ER, respectively.
The hollow portion of the ER tubules is called the lumen or cisternal space. The membrane of the ER, which is a phospholipid bilayer embedded with proteins, is continuous with the nuclear envelope.
Rough ER
The rough endoplasmic reticulum (RER) is so named because the ribosomes attached to its cytoplasmic surface give it a studded appearance when viewed through an electron microscope (see figure below).
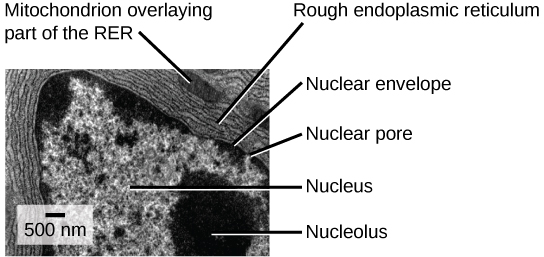
This transmission electron micrograph shows the rough endoplasmic reticulum and other organelles in a pancreatic cell. (credit: modification of work by Louisa Howard)
Ribosomes transfer their newly synthesized proteins into the lumen of the RER where they undergo structural modifications, such as folding or the acquisition of side chains. These modified proteins will be incorporated into cellular membranes—the membrane of the ER or those of other organelles—or secreted from the cell (such as protein hormones, enzymes). The RER also makes phospholipids for cellular membranes.
If the phospholipids or modified proteins are not destined to stay in the RER, they will reach their destinations via transport vesicles that bud from the RER’s membrane.
Since the RER is engaged in modifying proteins (such as enzymes, for example) that will be secreted from the cell, you would be correct in assuming that the RER is abundant in cells that secrete proteins. This is the case with cells of the liver, for example.
Smooth ER
The smooth endoplasmic reticulum (SER) is continuous with the RER but has few or no ribosomes on its cytoplasmic surface. Functions of the SER include synthesis of carbohydrates, lipids, and steroid hormones; detoxification of medications and poisons; and storage of calcium ions.
In muscle cells, a specialized SER called the sarcoplasmic reticulum is responsible for storage of the calcium ions that are needed to trigger the coordinated contractions of the muscle cells.
The Golgi Apparatus
We have already mentioned that vesicles can bud from the ER and transport their contents elsewhere, but where do the vesicles go? Before reaching their final destination, the lipids or proteins within the transport vesicles still need to be sorted, packaged, and tagged so that they wind up in the right place. Sorting, tagging, packaging, and distribution of lipids and proteins takes place in the Golgi apparatus (also called the Golgi body), a series of flattened membranes (see figure below).
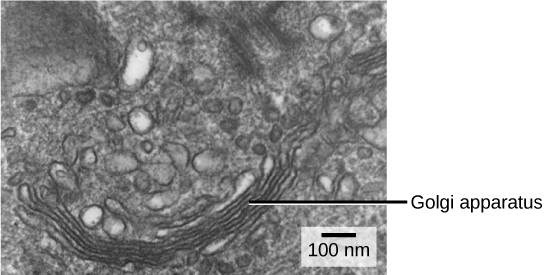
The Golgi apparatus in this white blood cell is visible as a stack of semicircular, flattened rings in the lower portion of the image. Several vesicles can be seen near the Golgi apparatus. (credit: modification of work by Louisa Howard)
The receiving side of the Golgi apparatus is called the cis face. The opposite side is called the trans face. The transport vesicles that formed from the ER travel to the cis face, fuse with it, and empty their contents into the lumen of the Golgi apparatus. As the proteins and lipids travel through the Golgi, they undergo further modifications that allow them to be sorted. The most frequent modification is the addition of short chains of sugar molecules. These newly modified proteins and lipids are then tagged with phosphate groups or other small molecules so that they can be routed to their proper destinations.
Finally, the modified and tagged proteins are packaged into secretory vesicles that bud from the trans face of the Golgi. While some of these vesicles deposit their contents into other parts of the cell where they will be used, other secretory vesicles fuse with the plasma membrane and release their contents outside the cell.
In another example of form following function, cells that engage in a great deal of secretory activity (such as cells of the salivary glands that secrete digestive enzymes or cells of the immune system that secrete antibodies) have an abundance of Golgi.
In plant cells, the Golgi apparatus has the additional role of synthesizing polysaccharides, some of which are incorporated into the cell wall and some of which are used in other parts of the cell.
Lysosomes
In addition to their role as the digestive component and organelle-recycling facility of animal cells, lysosomes are considered to be parts of the endomembrane system. Lysosomes also use their hydrolytic enzymes to destroy pathogens (disease-causing organisms) that might enter the cell. A good example of this occurs in a group of white blood cells called macrophages, which are part of your body’s immune system. In a process known as phagocytosis or endocytosis, a section of the plasma membrane of the macrophage invaginates (folds in) and engulfs a pathogen. The invaginated section, with the pathogen inside, then pinches itself off from the plasma membrane and becomes a vesicle. The vesicle fuses with a lysosome. The lysosome’s hydrolytic enzymes then destroy the pathogen (figure below).
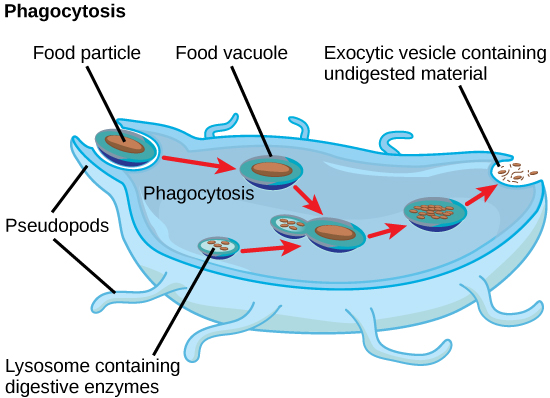
A macrophage has engulfed (phagocytized) a potentially pathogenic bacterium and then fuses with a lysosomes within the cell to destroy the pathogen. Other organelles are present in the cell but for simplicity are not shown.
Summary of Endomembranes
The endomembrane system includes the nuclear envelope, lysosomes, vesicles, the ER, and Golgi apparatus, as well as the plasma membrane. These cellular components work together to modify, package, tag, and transport proteins and lipids that form the membranes.
The RER modifies proteins and synthesizes phospholipids used in cell membranes. The SER synthesizes carbohydrates, lipids, and steroid hormones; engages in the detoxification of medications and poisons; and stores calcium ions. Sorting, tagging, packaging, and distribution of lipids and proteins take place in the Golgi apparatus. Lysosomes are created by the budding of the membranes of the RER and Golgi. Lysosomes digest macromolecules, recycle worn-out organelles, and destroy pathogens.
Free Response
Exercise 1
In the context of cell biology, what do we mean by form follows function? What are at least two examples of this concept?
“Form follows function” refers to the idea that the function of a body part dictates the form of that body part. As an example, compare your arm to a bat’s wing. While the bones of the two correspond, the parts serve different functions in each organism and their forms have adapted to follow that function.
Exercise 2
In your opinion, is the nuclear membrane part of the endomembrane system? Why or why not? Defend your answer.
Since the external surface of the nuclear membrane is continuous with the rough endoplasmic reticulum, which is part of the endomembrane system, then it is correct to say that it is part of the system.
The cytoskeleton
Section summary
The cytoskeleton is a network of different protein fibers that provides many functions: it maintains or changes the shape of the cell; it secures some organelles in specific positions; it enables movement of cytoplasm and vesicles within the cell; and it enables the cell to move in response to stimuli. There are three types of fibers within the cytoskeleton: microfilaments, intermediate filaments, and microtubules. Some of the cytoskeletal fibers work in conjunction with molecular motors which move along the fibers within the cell to carry out a diverse set of functions. There are two main families of cytoskeletally-associated molecular motors: dyneines and kinesins.
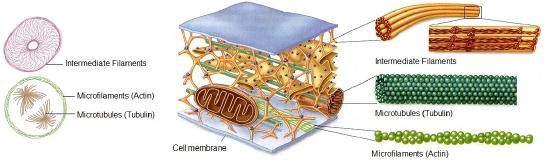
Figure 1. Microfilaments thicken the cortex around the inner edge of a cell; like rubber bands, they resist tension. Microtubules are found in the interior of the cell where they maintain cell shape by resisting compressive forces. Intermediate filaments are found throughout the cell and hold organelles in place.
Design challenge
Problem statement: Eukaryotic cells contain membrane-bound organelles that effectively separate materials, processes, and reactions from one another and from the cytoplasm. This in itself poses a problem for eukaryotes.
How can the cell purposely move and control the location of materials between these organelles? More specifically, how can a eukaryotic cell transport compounds from their place of origin (in most cases the cyotoplasm) to where they are needed (perhaps the nucleus, the mitochondria, or the cell surface)?
Note: possible discussion
Propose some reasons why cells—particularly large cells and/or cells with organelles—cannot rely on simple diffusion to move metabolites, building blocks, proteins, etc. to the locations in the cell where they are needed.
One possible solution is for the cell to create a network that can connect all the different parts of the cell together. This network could be used not only as a scaffold to hold components in place but also as a reference for direction. For example, we can use a map to determine the direction we need to travel and roads to connect and travel from home to campus. Likewise, an interconnecting network inside the cell can be used to direct and move compounds from one location to a final destination. Some of the required characteristics of this network are listed below. Can you add to this list?
Intracellular network
- The network needs to be extensive, and connect every area of the cell.
- The network needs to be flexible, able to change and adapt as the cell grows larger, divides into two cells, or physically moves from one environment to another.
- The network needs to be strong, able to hold up to mechanical pressure from inside the cell or from outside of the cell.
- The network needs to be composed of different fibers and each of these fibers needs to be for a specific connection in the cell. For example, certain fibers might be involved in holding organelles in place, and other fibers would be involved in connecting two different organelles.
- The fibers need to have directionality (or polarity), meaning they need to have a defined starting point and a defined end to help direct movement from one location to another.
- The fibers need to work with proteins that can convert chemical energy into kinetic energy, to actively transport compounds along the fibers.
Microfilaments
Actin
Microfilaments are cytoskeleton fibers composed of actin subunits. Actin is one of the most abundant proteins in eukaryotic cells and comprises 20% of total cellular protein by weight in muscle cells. The actin amino acid sequence is highly conserved in eukaryotic cells, meaning that the protein amino acid sequence, and therefore its final 3-D shape, has changed little over the course of evolution, maintaining more than 80% similarity between algae and humans.
Actin can be present as either a free monomer called G-actin (globular) or as part of a polymer microfilament called F-actin ("F" for filamentous). Actin must be bound to ATP in order to assemble into its filamentous form and maintain the structural integrity of the filament. The actin filament itself has structural polarity. This term "polarity", in reference to a cytoskeleton filament, does not mean what it did when we discussed polar functional groups earlier in this course. Polarity here refers to the fact that there are two distinct ends to the filament. These ends are called the "(-)" end and the "(+)" end. At the "(+)" end, actin subunits are adding onto the elongating filament and at the "(-)" end, actin subunits are disassembling or falling off of the filament. This process of assembly and disassembly is controlled by the ATP to ADP ratio in the cytoplasm.
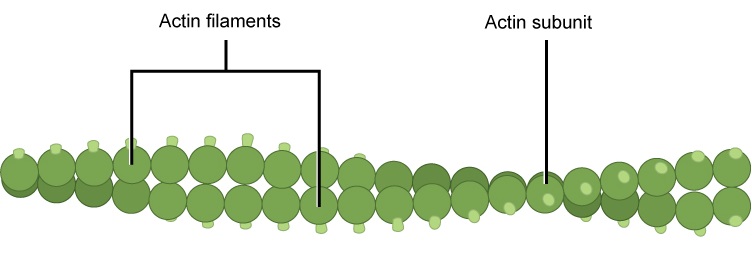
Figure 2. Microfilaments are the narrowest of the three cytoskeleton fibers, with a diameter of about seven nm. Microfilaments are composed of actin subunits which form into two intertwined strands.
Actin participates in many cellular processes, including muscle contraction, cell motility, cytokinesis during cell division, vesicle and organelle movement, and the maintenance of cell shape. Actin filaments serve as a track for the movement of a family of motor proteins called myosins discussed in more detail in a section below.
Link to learning:
To see an example of a white blood cell in action, click here and watch a short time-lapse video of the cell capturing two bacteria. It engulfs one and then moves on to the other.
Animations on actin filaments and how they work
Intermediate filaments
Intermediate filaments are made of several strands of fibrous proteins that are wound together. These elements of the cytoskeleton get their name from the fact that their diameter, eight to ten nm, is between those of the smaller microfilaments and the larger microtubules. The intermediate filaments are the most diverse group of cytoskeletal elements. Several types of fibrous proteins are found in the intermediate filaments. You are probably most familiar with keratin, the fibrous protein that strengthens your hair, nails, and the epidermis of the skin.
Figure 3. Intermediate filaments consist of several intertwined strands of fibrous proteins.
Intermediate filaments have no role in cell movement. Their function is purely structural. They bear tension, thus maintaining the shape of the cell, and anchor the nucleus and other organelles in place. The figure above shows how intermediate filaments create a cable-like supportive scaffolding inside the cell.
Microtubules
Microtubules are the largest component of the cytoskeleton and are found throughout the cytoplasm. These polymers are made up of globular protein subunits called α-tubulin and β-tubulin. Microtubules are found not only in eukaryotic cells but in some bacteria as well.
Both the α-tubulin and β-tubulin subunits bind to GTP. When bound to GTP, the formation of the microtubule can begin, this is called the nucleation event. As more GTP tubulin dimers assemble onto the filament, GTP is slowly hydrolyzed by β-tubulin to form GDP. Tubulin bound to GDP is less structurally robust and can lead to disassembly of the microtubule.
Much like the actin filaments discussed above, microtubules also have a distinct polarity that is critical for their biological function. Tubulin polymerizes end to end, with the β-subunits of one tubulin dimer contacting the α-subunits of the next dimer. These differences lead to different subunits being exposed on the two ends of the filament. The ends are designated the "(−)" and "(+)" ends. Unlike actin filaments, microtubules can elongate at both the "(+)" and "(-)" ends, but elongation is significantly more rapid at the "(+)" end.
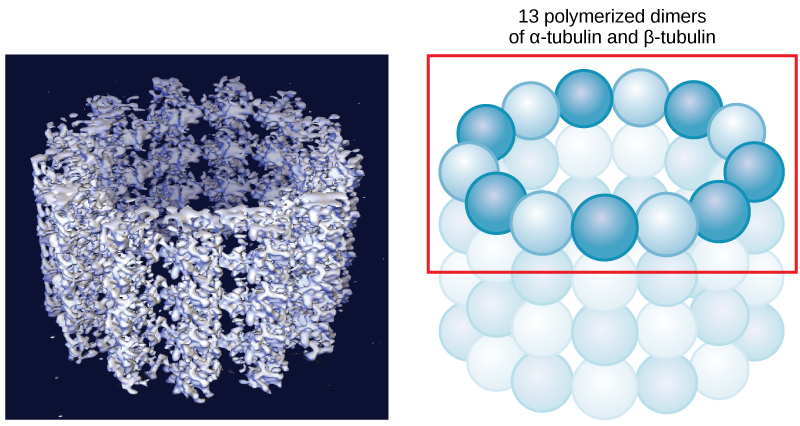
Figure 4. Microtubules are hollow. Their walls consist of 13 polymerized dimers of α-tubulin and β-tubulin (right image). The left image shows the molecular structure of the tube.
Microtubules help the cell resist compression, provide a track along which vesicles move through the cell, pull replicated chromosomes to opposite ends of a dividing cell, and are the structural elements of flagella, cilia, and centrioles (the latter are the two perpendicular bodies of the centrosome). In fact, in animal cells, the centrosome is the microtubule organizing center. In eukaryotic cells, flagella and cilia are quite different structurally from their counterparts in bacteria, discussed below.
Animations of the cytoskeleton
Where did these fibers come from?
The cytoskeleton probably has its origins in bacterial and/or archaeal ancestry. There are ancient relatives to both actin and tubulin in bacterial systems. In bacteria, the MreB protein and the ParM protein are believed to be early ancestors to Actin. MreB functions in maintaining cell shape and ParM functions in plasmid (DNA) partitioning. The FtsZ protein in bacteria functions in cytokinesis, it is a GTPase, spontaneously forms filaments and is hypothesized to be an ancient form of tubulin. These findings support the hypothesis that the eukaryotic cytoskeleton has its origins in the bacterial world.
Flagella and cilia
Flagella (singular=flagellum) are long, hair-like structures that extend from the plasma membrane and are used to move an entire cell (for example, sperm, Euglena). When present, the cell has just one flagellum or a few flagella. Cilia are short, hair-like structures that are used to move entire cells (such as paramecia) or substances along the outer surface of the cell (for example, the cilia of cells lining the fallopian tubes that move the ovum toward the uterus, or cilia lining the cells of the respiratory tract that trap particulate matter and move it toward your nostrils.) When cilia are present, there can be many of them, extending along the entire surface of the plasma membrane.
Despite their differences in length and number, flagella and cilia share a common structural arrangement of microtubules called a “9+2 array.” This is an appropriate name because a single flagellum or cilium is made of a ring of nine microtubule doublets, surrounding a single microtubule doublet in the center (Figure 5).
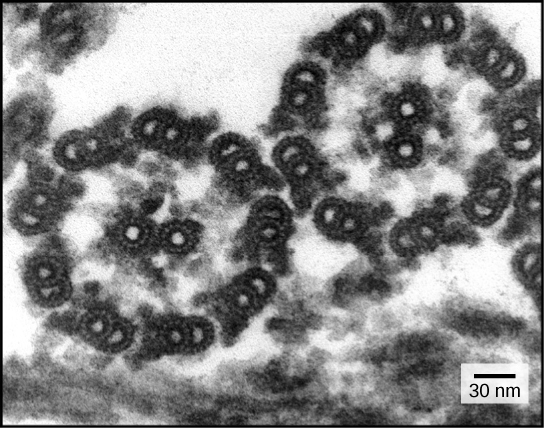
Figure 5. This transmission electron micrograph of two flagella shows the "9+2 array" of microtubules: nine microtubule doublets surround a single microtubule doublet. (credit: modification of work by Dartmouth Electron Microscope Facility, Dartmouth College; scale-bar data from Matt Russell)
For a video on flagellar and ciliar movement in eukaryotes, see the YouTube video: click here (you can skip the commericial).
Motor proteins
One function of the cytoskeleton is to move cellular components from one part of the cell to another. These cellular components are called "cargo" and are often stored within a vesicle for transport. You can think of the cytoskeleton as "railroad tracks" providing support and directionality inside of the cell.
Of course, if there are "railroad tracks" there needs to be an engine that can both move on the tracks and pull or push cargo along. In this case the engines are molecular motors that can move along the tracks in a specific direction. There are two families of molecular motors associated with the cytoskeleton; dyneines and kinesins. These motor proteins (train engines) and the cytoskeleton create a comprehensive network within the cell for moving vesicles (box cars) from one organelle to another or from one organelle to the cell surface.
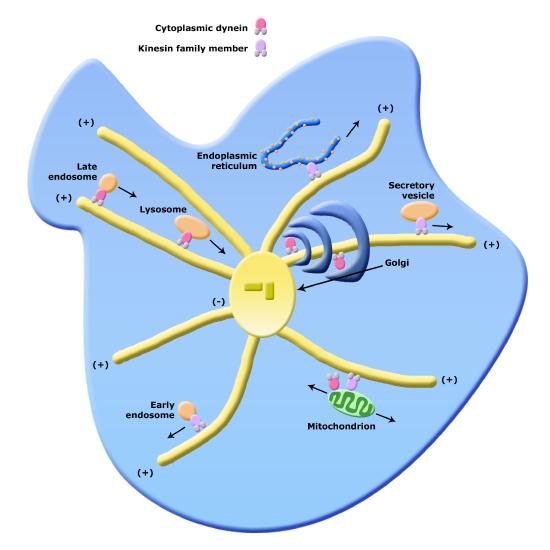
Figure 6. Organelle transport via microtubules and kinesins and dynes. Note that the figure is conceptual and only intended to show directionality of movement of various organelles; it does not necessarily represent all of their forms faithfully.
Cytoplasmic dyneins
Dynein is a protein complex that functions as a molecular motor. In cells, it converts the chemical energy from ATP hydrolysis into the mechanical energy of movement to 'walk' along the microtubule while carrying a vesicle. Dyneins bind to microtubules and move or "walk" from the plus "(+)" end of the cytoskeletal microtubule filament to the minus "(-)" end of the filament, which is usually oriented towards the cell center. Thus, they are often referred to as "minus end directed motors" and this vesicular transport is refereed to as retrograde transport. Cytoplasmic dynein moves processively along the microtubule, hydrolyzing ATP with each "step" it takes along the microtubule. During this process, one or the other of its "stalks" is always attached to the microtubule, allowing for the dynein motor (and its cargo) to "walk" a considerable distance along a microtubule without detaching.
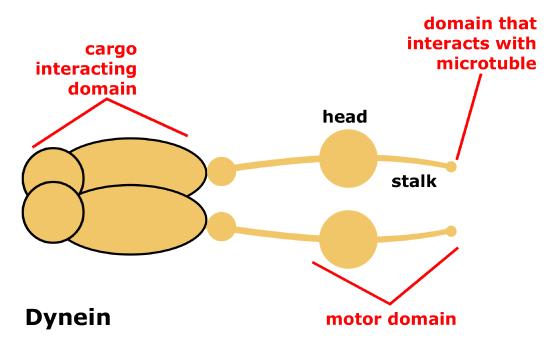
Figure 7. Schematic of cytoplasmic dynein motor protein. Dyneins are protein complexes composed of many smaller polypeptide subunits. The overall structure of the dynien motors are relatively simple, consisting of two identical complexes each having a motor domain that interacts with the microtubule, a stalk, or stem region that connects the motor head to the cargo interacting domain.
Cytoplamic dyneins are used in many different processes: they are involved in organelle movement such as the positioning of the Golgi complex and other organelles in the cell; they are used in the transport of cargo such as the movement of vesicles made by the endoplasmic reticulum, endosomes, and lysosomes; and they are responsible for the movement of chromosomes during cell division. Axonemal dyneins are motor proteins used in the sliding of microtubules in the axonemes of cilia and flagella in eukaryotic cells.
Kinesins
Kinesins, like cytoplasmic dyneins are motor-protein complexes that "walk" along the microtubules and are involved in vesicle transport. Unlike cytoplasmic dyneins, the polarity of kinesin movement is from the "(-)" end of the microtubule to the "(+)" end with the hydrolysis of ATP. In most cells, this entails transporting cargo from the center of the cell towards the periphery (the opposite direction to dyneins). This form of transport is known as anterograde or orthrograde transport. Like cytoplasmic dyneins, kinesins are involved in a variety of cellular processes including vesicle movement and chromosome movement during cell division.
The structure of kinesins are similar to cytoplasmic dyneins and is diagrammed in Figure 8. Members of the kinesin superfamily vary in shape, but the overall structure is that of a heterotetramer whose motor subunits (heavy chains) form a protein dimer (molecule pair) that binds two light chains.
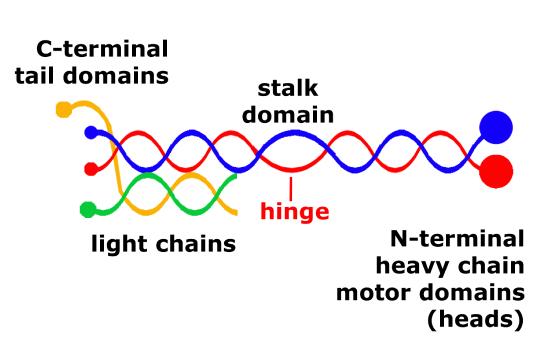
Figure 8. Schematic of kinesin motor proteins. The heavy chains comprise a globular head (the motor domain) at the amino terminal end connected via a short, flexible neck linker to the stalk—a long, central α-helical coiled-coil domain—that ends in a carboxy terminal tail domain which associates with the light-chains. The stalks of two light chains intertwine to form a coiled-coil that directs dimerization of the two heavy chains. In most cases transported cargo binds to the kinesin light chains, but in some cases cargo binds to the C-terminal domains of the heavy chains.
Animations of kinesin and dynein at work
How do the motors interact with cargo and the microtubules?
Cytoplasmic dyneins and kinesins interact with both cargo and microtubules in similar fashion. The light chains interact with receptors on the various cargo vesicles and the globular motor domains, specifically interact with the microtubules.
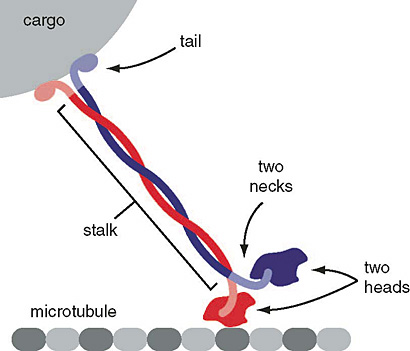
Figure 9. Schematic of kinesin motor protein carrying a cargo vesicle along a microtubule filament.
Note: possible discussion
What are the benefits for having multiple types of motor proteins? Multiple types of filaments? Filaments with polarity?