W2018_Bis2A_Lecture16_reading
- Page ID
- 25328
Light Energy and Pigments
Light Energy
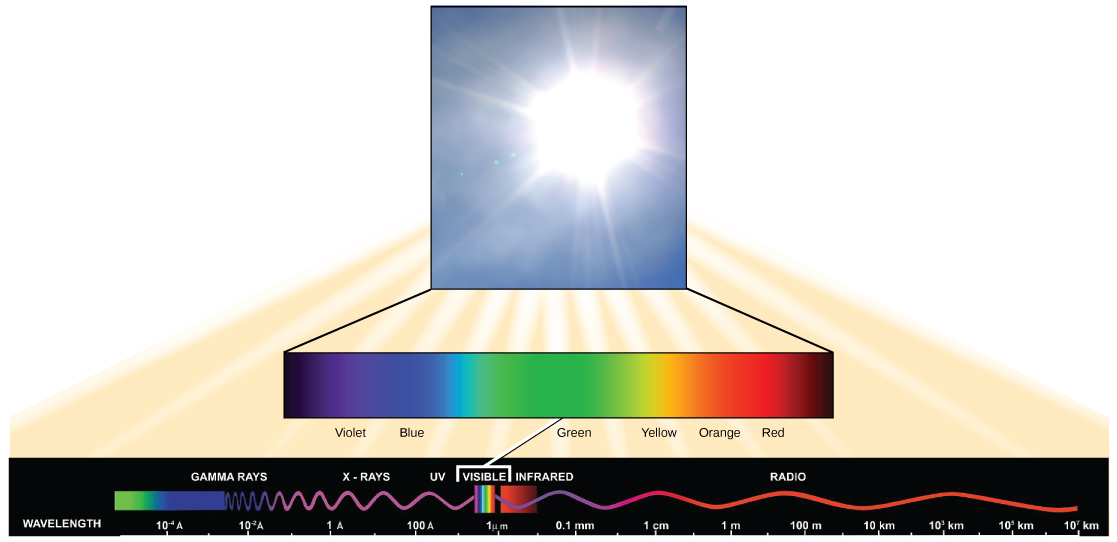
The sun emits an enormous amount of electromagnetic radiation (solar energy) that spans a broad swath of the electromagnetic spectrum, the range of all possible radiation frequencies. When solar radiation reaches Earth, a fraction of this energy interacts with and may be transferred to the matter on the planet. This energy transfer results in a wide variety of different phenomena, from influencing weather patterns to driving a myriad of biological processes. In BIS2A, we are largely concerned with the latter, and below, we discuss some very basic concepts related to light and its interaction with biology.
First, however we need to refresh a couple of key properties of light:
- Light in a vacuum travels at a constant speed of 299,792,458 m/s. We often abbreviate the speed of light with the variable "c".
- Light has properties of waves. A specific "color" of light has a characteristic wavelength.

The distance between peaks in a wave is referred to as the wavelength and is abbreviated with the greek letter lambda (Ⲗ).
Attribution: Marc T. Facciotti (original work)
The inverse proportionality of frequency and wavelength. Wave 1 has a wavelength that is 2x that of wave 2 (Ⲗ1 > Ⲗ2). If the two waves are traveling at the same speed (c)—imagine that both of the whole lines that are drawn are dragged past the fixed vertical line at the same speed —then the number of times a wave peak passes a fixed point is greater for wave 2 than wave 1 (f2 > f1).
Attribution: Marc T. Facciotti (original work)
3. Finally, each frequency (or wavelength) of light is associated with a specific energy. We'll call energy "E". The relationship between frequency and energy is:
E = h*f
where h is a constant called the Planck constant (~6.626x10-34 Joule•second when frequency is expressed in cycles per second). Given the relationship between frequency and wavelength, you can also write E = h*c/Ⲗ. Therefore, the larger the frequency (or shorter the wavelength), the more energy is associated with a specific "color". Wave 2 in the figure above is associated with greater energy than wave 1.
The sun emits energy in the form of electromagnetic radiation. All electromagnetic radiation, including visible light, is characterized by its wavelength. The longer the wavelength, the less energy it carries. The shorter the wavelength, the more energy is associated with that band of the electromagnetic spectrum.
The Light We See
The visible light seen by humans as white light is composed of a rainbow of colors, each with a characteristic wavelength. Certain objects, such as a prism or a drop of water, disperse white light to reveal the colors to the human eye. In the visible spectrum, violet and blue light have shorter (higher energy) wavelengths while the orange and red light have longer (lower energy) wavelengths.
The colors of visible light do not carry the same amount of energy. Violet has the shortest wavelength and therefore carries the most energy, whereas red has the longest wavelength and carries the least amount of energy.
Credit: modification of work by NASA
Absorption by Pigments
The interaction between light and biological systems occurs through several different mechanisms, some of which you may learn about in upper division courses in cellular physiology or biophysical chemistry. In BIS2A, we are mostly concerned with the interaction of light and biological pigments. These interactions can initiate a variety of light-dependent biological processes that can be grossly grouped into two functional categories: cellular signaling and energy harvesting. Signaling interactions are largely responsible for perceiving changes in the environment (in this case, changes in light). An example of a signaling interaction might be the interaction between light and the pigments expressed in an eye. By contrast, light/pigment interactions that are involved in energy harvesting are used for—not surprisingly—capturing the energy in the light and transferring it to the cell to fuel biological processes. Photosynthesis, which we will learn more about soon, is one example of an energy harvesting interaction.
At the center of the biological interactions with light are groups of molecules we call organic pigments. Whether in the human retina, chloroplast thylakoid, or microbial membrane, organic pigments often have specific ranges of energy or wavelengths that they can absorb. The sensitivity of these molecules for different wavelengths of light is due to their unique chemical makeups and structures. A range of the electromagnetic spectrum is given a couple of special names because of the sensitivity of some key biological pigments: The retinal pigment in our eyes, when coupled with an opsin sensor protein, “sees” (absorbs) light predominantly between the wavelengths between of 700 nm and 400 nm. Because this range defines the physical limits of the electromagnetic spectrum that we can actually see with our eyes, we refer to this wavelength range as the "visible range". For similar reasons, as plants pigment molecules tend to absorb wavelengths of light mostly between 700 nm and 400 nm, plant physiologists refer to this range of wavelengths as "photosynthetically active radiation".
Three Key Types of Pigments We Discuss in BIS2A
Chlorophylls
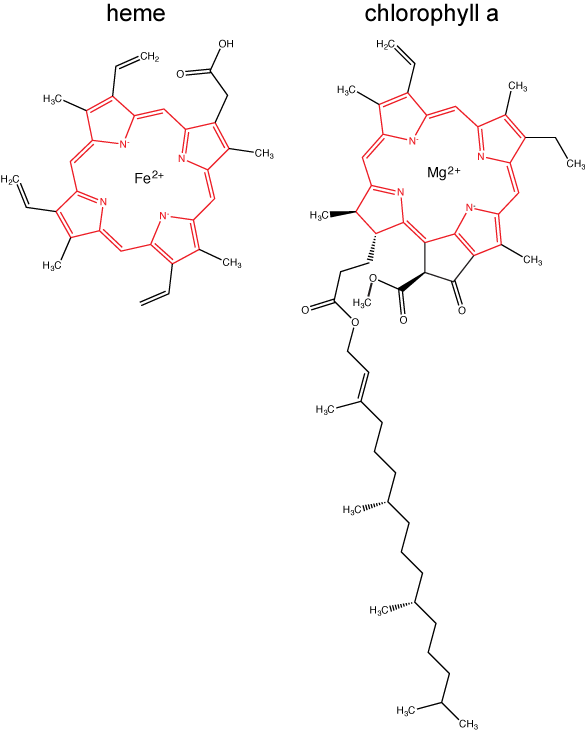
Chlorophylls (including bacteriochlorophylls) are part of a large family of pigment molecules. There are five major chlorophyll pigments named: a, b, c, d, and f. Chlorophyll a is related to a class of more ancient molecules found in bacteria called bacteriochlorophylls. Chlorophylls are structurally characterized by ring-like porphyrin group that coordinates a metal ion. This ring structure is chemically related to the structure of heme compounds that also coordinate a metal and are involved in oxygen binding and/or transport in many organisms. Different chlorophylls are distinguished from one another by different "decorations"/chemical groups on the porphyrin ring.
The structure of heme and chlorophyll a molecules. The common porphyrin ring is colored in red.
Attribution: Marc T. Facciotti (original work)
Carotenoids
Carotenoids are the red/orange/yellow pigments found in nature. They are found in fruit—the red of tomato (lycopene), the yellow of corn seeds (zeaxanthin), or the orange of an orange peel (β-carotene)—which are used as biological "advertisements" to attract seed dispersers (animals or insects that may carry seeds elsewhere). In photosynthesis, carotenoids function as photosynthetic pigments. In addition, when a leaf is exposed to full sun, that surface is required to process an enormous amount of energy; if that energy is not handled properly, it can do significant damage. Therefore, many carotenoids help absorb excess energy in light and safely dissipate that energy as heat.
Flavonoids
Flavonoids are a very broad class of compounds that are found in great diversity in plants. These molecules come in many forms but all share a common core structure shown below. The diversity of flavonoids comes from the many different combinations of functional groups that can "decorate" the core flavone.

The core ring structure of flavans.
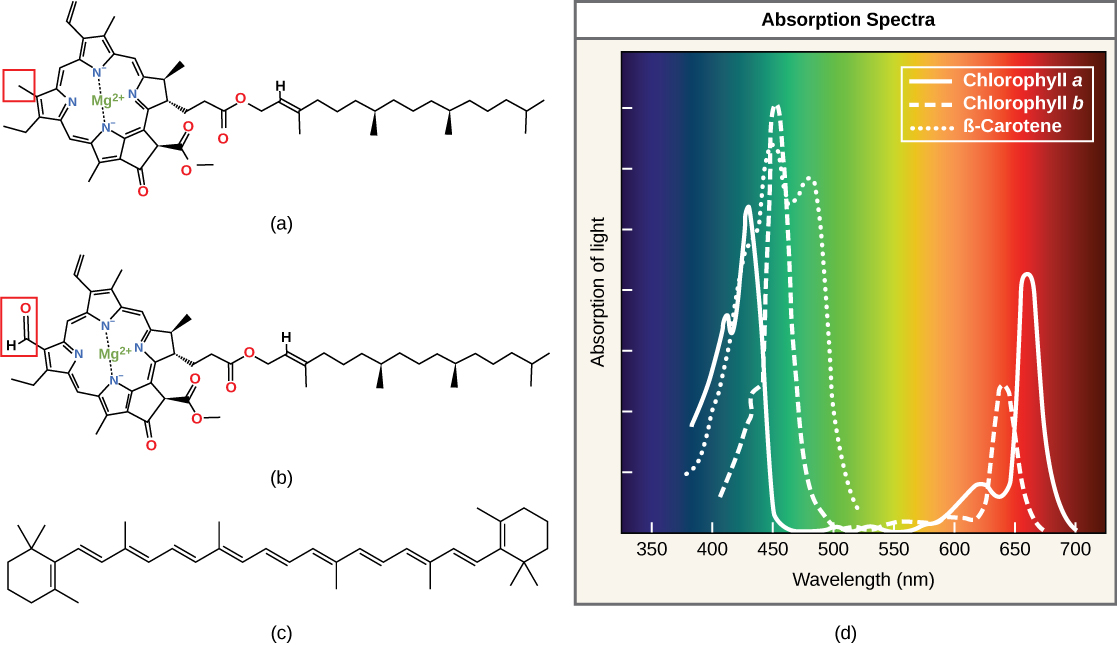
Each type of pigment can be identified by the specific pattern of wavelengths it absorbs from visible light. This characteristic is known as the pigment's absorption spectrum. The graph in the figure below shows the absorption spectra for chlorophyll a, chlorophyll b, and a type of carotenoid pigment called β-carotene (which absorbs blue and green light). Notice how each pigment has a distinct set of peaks and troughs, revealing a highly specific pattern of absorption. These differences in absorbance are due to differences in chemical structure (some are highlighted in the figure). Chlorophyll a absorbs wavelengths from either end of the visible spectrum (blue and red), but not green. Because green is reflected or transmitted, chlorophyll appears green. Carotenoids absorb in the short-wavelength blue region, and reflect the longer yellow, red, and orange wavelengths.
(a) Chlorophyll a, (b) chlorophyll b, and (c) β-carotene are hydrophobic organic pigments found in the thylakoid membrane. Chlorophyll a and b, which are identical except for the part indicated in the red box, are responsible for the green color of leaves. Note how the small amount of difference in chemical composition between different chlorophylls leads to different absorption spectra. β-carotene is responsible for the orange color in carrots. Each pigment has a unique absorbance spectrum (d).
Importance of having multiple different pigments
Not all photosynthetic organisms have full access to sunlight. Some organisms grow underwater where light intensity and available wavelengths decrease and change, respectively, with depth. Other organisms grow in competition for light. For instance, plants on the rainforest floor must be able to absorb any bit of light that comes through because the taller trees absorb most of the sunlight and scatter the remaining solar radiation. To account for these variable light conditions, many photosynthetic organisms have a mixture of pigments whose expression can be tuned to improve the organism's ability to absorb energy from a wider range of wavelengths than would be possible with one pigment alone.
Photophosphorylation
Photophosphorylation an overview
Photophosphorylation is the process of transferring the energy from light into chemicals, particularly ATP. The evolutionary roots of photophosphorylation are likely in the anaerobic world, between 3 billion and 1.5 billion years ago, when life was abundant in the absence of molecular oxygen. Photophosphorylation probably evolved relatively shortly after electron transport chains (ETC) and anaerobic respiration began to provide metabolic diversity. The first step of the process involves the absorption of a photon by a pigment molecule. Light energy is transferred to the pigment and promotes electrons (e-) into a higher quantum energy state—something biologists term an "excited state". Note the use of anthropomorphism here; the electrons are not "excited" in the classic sense and aren't all of a sudden hopping all over or celebrating their promotion. They are simply in a higher energy quantum state. In this state, the electrons are colloquially said to be "energized". While in the "excited" state, the pigment now has a much lower reduction potential and can donate the "excited" electrons to other carriers with greater reduction potentials. These electron acceptors may, in turn, become donors to other molecules with greater reduction potentials and, in doing so, form an electron transport chain.
As electrons pass from one electron carrier to another via red/ox reactions, these exergonic transfers can be coupled to the endergonic transport (or pumping) of protons across a membrane to create an electrochemical gradient. This electrochemical gradient generates a proton motive force whose exergonic drive to reach equilibrium can be coupled to the endergonic production of ATP, via ATP synthase. As we will see in more detail, the electrons involved in this electron transport chain can have one of two fates: (1) they may be returned to their initial source in a process called cyclic photophosphorylation; or (2) they can be deposited onto a close relative of NAD+ called NADP+. If the electrons are deposited back on the original pigment in a cyclic process, the whole process can start over. If, however, the electron is deposited onto NADP+ to form NADPH (**shortcut note—we didn't explicitly mention any protons but assume it is understood that they are also involved**), the original pigment must regain an electron from somewhere else. This electron must come from a source with a smaller reduction potential than the oxidized pigment and depending on the system there are different possible sources, including H2O, reduced sulfur compounds such as SH2 and even elemental S0.
What happens when a compound absorbs a photon of light?
When a compound absorbs a photon of light, the compound is said to leave its ground state and become "excited".
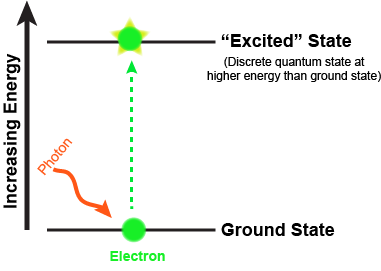
What are the fates of the "excited" electron? There are four possible outcomes, which are schematically diagrammed in the figure below. These options are:
- The e- can relax to a lower quantum state, transferring energy as heat.
- The e- can relax to a lower quantum state and transfer energy into a photon of light—a process known as fluorescence.
- The energy can be transferred by resonance to a neighboring molecule as the e- returns to a lower quantum state.
- The energy can change the reduction potential such that the molecule can become an e- donor. Linking this excited e- donor to a proper e- acceptor can lead to an exergonic electron transfer. In other words, the excited state can be involved in red/ox reactions.
As the excited electron decays back to its lower energy state, the energy can be transferred in a variety of ways. While many so called antenna or auxiliary pigments absorb light energy and transfer it to something known as a reaction center (by mechanisms depicted in option III in Figure 2), it is what happens at the reaction center that we are most concerned with (option IV in the figure above). Here a chlorophyll or bacteriochlorophyll molecule absorbs a photon's energy and an electron is excited. This energy transfer is sufficient to allow the reaction center to donate the electron in a red/ox reaction to a second molecule. This initiates the electron transport reactions. The result is an oxidized reaction center that must now be reduced in order to start the process again. How this happens is the basis of electron flow in photophosphorylation and will be described in detail below.
Simple photophosphorylation systems: anoxygenic photophosphorylation
Early in the evolution of photophosphorylation, these reactions evolved in anaerobic environments where there was very little molecular oxygen available. Two sets of reactions evolved under these conditions, both directly from anaerobic respiratory chains as described previously. These are known as the light reactions because they require the activation of an electron (an "excited" electron) from the absorption of a photon of light by a reaction center pigment, such as bacteriochlorophyll. The light reactions are categorized either as cyclic or as noncyclic photophosphorylation, depending upon the final state of the electron(s) removed from the reaction center pigments. If the electron(s) returns to the original pigment reaction center, such as bacteriochlorophyll, this is cyclic photophosphorylation; the electrons make a complete circuit and is diagramed in Figure 4. If the electron(s) are used to reduce NADP+ to NADPH, the electron(s) are removed from the pathway and end up on NADPH; this process is referred to as noncyclic since the electrons are no longer part of the circuit. In this case the reaction center must be re-reduced before the process can happen again. Therefore, an external electron source is required for noncylic photophosphorylation. In these systems reduced forms of Sulfur, such as H2S, which can be used as an electron donor and is diagrammed in Figure 5. To help you better understand the similarities of photophosphorylation to respiration, a red/ox tower has been provided that contains many commonly used compounds involved with photosphosphorylation.
|
oxidized form |
reduced form |
n (electrons) |
Eo´ (volts) |
|---|---|---|---|
|
PS1* (ox) |
PS1* (red) |
- |
-1.20 |
|
ferredoxin (ox) version 1 |
ferredoxin (red) version 1 |
1 |
-0.7 |
|
PSII* (ox) |
PSII* (red) |
- |
-0.67 |
|
P840* (ox) |
PS840* (red) |
- |
-0.67 |
|
acetate |
acetaldehyde |
2 |
-0.6 |
|
CO2 |
Glucose |
24 |
-0.43 |
|
ferredoxin (ox) version 2 |
ferredoxin (red) version 2 |
1 |
-0.43 |
|
CO2 |
formate |
2 |
-0.42 |
|
2H+ |
H2 |
2 |
-0.42 |
|
NAD+ + 2H+ |
NADH + H+ |
2 |
-0.32 |
|
NADP+ + 2H+ |
NADPH + H+ |
2 |
-0.32 |
|
Complex I FMN (enzyme bound) |
FMNH2 |
2 |
-0.3 |
|
Lipoic acid, (ox) |
Lipoic acid, (red) |
2 |
-0.29 |
|
FAD+ (free) + 2H+ |
FADH2 |
2 |
-0.22 |
|
Pyruvate + 2H+ |
lactate |
2 |
-0.19 |
|
FAD+ + 2H+ (bound) |
FADH2 (bound) |
2 |
0.003-0.09 |
|
CoQ (Ubiquinone - UQ + H+) |
UQH. |
1 |
0.031 |
|
UQ + 2H+ |
UQH2 |
2 |
0.06 |
|
Plastoquinone; (ox) |
Plastoquinone; (red) |
- |
0.08 |
|
Ubiquinone; (ox) |
Ubiquinone; (red) |
2 |
0.1 |
|
Complex III Cytochrome b2; Fe3+ |
Cytochrome b2; Fe2+ |
1 |
0.12 |
|
Complex III Cytochrome c1; Fe3+ |
Cytochrome c1; Fe2+ |
1 |
0.22 |
|
Cytochrome c; Fe3+ |
Cytochrome c; Fe2+ |
1 |
0.25 |
|
Complex IV Cytochrome a; Fe3+ |
Cytochrome a; Fe2+ |
1 |
0.29 |
|
1/2 O2 + H2O |
H2O2 |
2 |
0.3 |
|
P840GS (ox) |
PS840GS (red) |
- |
0.33 |
|
Complex IV Cytochrome a3; Fe3+ |
Cytochrome a3; Fe2+ |
1 |
0.35 |
|
Ferricyanide |
ferrocyanide |
2 |
0.36 |
|
Cytochrome f; Fe3+ |
Cytochrome f; Fe2+ |
1 |
0.37 |
|
PSIGS (ox) |
PSIGS (red) |
. |
0.37 |
|
Nitrate |
nitrite |
1 |
0.42 |
|
Fe3+ |
Fe2+ |
1 |
0.77 |
|
1/2 O2 + 2H+ |
H2O |
2 |
0.816 |
|
PSIIGS (ox) |
PSIIGS (red) |
- |
1.10 |
|
* Excited State, after absorbing a photon of light GS Ground State, state prior to absorbing a photon of light PS1: Oxygenic photosystem I P840: Bacterial reaction center containing bacteriochlorophyll (anoxygenic) PSII: Oxygenic photosystem II |
|||
Cyclic photophosphorylation
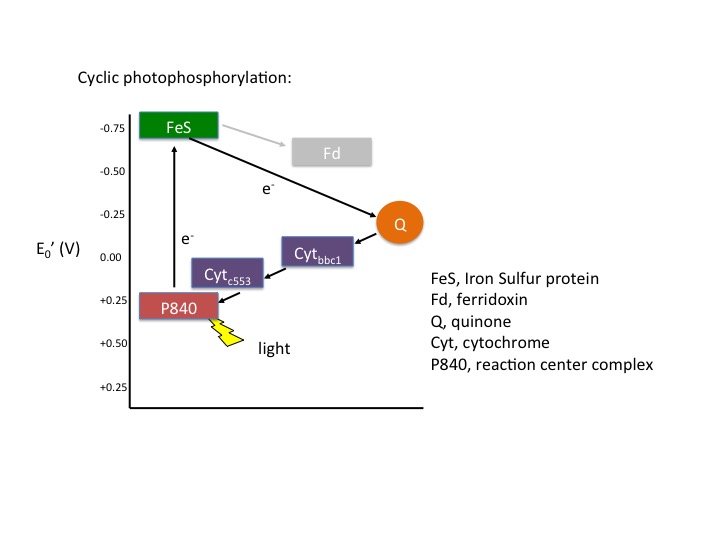
In cyclic photophosphorylation the bacteriochlorophyllred molecule absorbs enough light energy to energize and eject an electron to form bacteriochlorophyllox. The electron reduces a carrier molecule in the reaction center which in turn reduces a series of carriers via red/ox reactions. These carriers are the same carriers found in respiration. If the change in reduction potential from the various red/ox reactions are sufficiently large, H+ protons can be translocated across a membrane. Eventually, the electron is used to reduce bacteriochlorophyllox (making a complete loop) and the whole process can start again. This flow of electrons is cyclic and is therefore said to drive a processed called cyclic photophosphorylation. The electrons make a complete cycle: bacteriochlorophyll is the initial source of electrons and is the final electron acceptor. ATP is produced via the F1F0 ATPase. The schematic in Figure 4 demonstrates how cyclic electrons flow and thus how cyclic photophosphorylation works.
Figure 4. Cyclic electron flow. The reaction center P840 absorbs light energy and becomes excited, denoted with an *. The excited electron is ejected and used to reduce an FeS protein leaving an oxidized reaction center. The electron its transferred to a quinone, then to a series of cytochromes, which in turn reduces the P840 reaction center. The process is cyclical. Note the gray array coming from the FeS protein going to a ferridoxin (Fd), also in gray. This represents an alternative pathway the electron can take and will be discussed below in noncyclic photophosphorylation. Note that the electron that initially leaves the P840 reaction center is not necessarily the same electron that eventually finds its way back to reduce the oxidized P840.
Note: possible discussion
The figure of cyclic photophosphorylation above depicts the flow of electrons in a respiratory chain. How does this process help generate ATP?
Noncyclic photophosphorylation
In cyclic photophosphorylation, electrons cycle from bacteriochlorophyll (or chlorophyll) to a series of electron carriers and eventually back to bacteriochlorophyll (or chlorophyll); there is theoretically no net loss of electrons and they stay in the system. In noncyclic photophosphorylation, electrons are removed from the photosystem and red/ox chain and eventually end up on NADPH. That means there needs to be a source of electrons, a source that has a smaller reduction potential than bacteriochlorophyll (or chlorophyll) that can donate electrons to bacteriochlorophyllox to reduce it. From looking at the electron tower in Figure 3, you can see what compounds can be used to reduce the oxidized form of bacteriochlorophyll. The second requirement is that, when bacteriochlorophyll becomes oxidized and the electron is ejected, it must reduce a carrier that has a greater reduction potential than NADP/NADPH (see the electron tower). In this case, electrons can flow from energized bacteriochlorophyll to NADP forming NADPH and oxidized bacteriochlorophyll. Electrons are lost from the system and end up on NADPH; to complete the circuit, bacteriochlorophyllox is reduced by an external electron donor such as H2S or elemental S0.
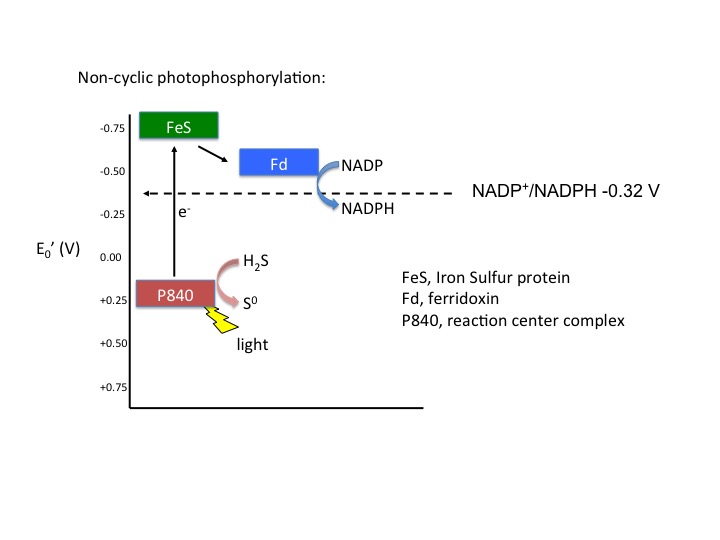
Noncyclic electron flow
Figure 5. Noncyclic electron flow. In this example, the P840 reaction center absorbs light energy and becomes energized; the emitted electron reduces a FeS protein and in turn reduces ferridoxin. Reduced ferridoxin (Fdred) can now reduce NADP to form NADPH. The electrons are now removed from the system, finding their way to NADPH. The electrons need to be replaced on P840, which requires an external electron donor. In this case, H2S serves as the electron donor.
Note: possible discussion
It should be noted that for bacterial photophosphorylation pathways, for each electron donated from a reaction center [remember only one electron is actually donated to the reaction center (or chlorophyl molecule)], the resulting output from that electron transport chain is either the formation of NADPH (requires two electrons) or ATP can be made but NOT not both. In other words, the path the electrons take in the ETC can have one or two possible outcomes. This puts limits on the versatility of the bacterial anoxygenic photosynthetic systems. But what would happen if there evolved a process that utilized both systems, that is, a cyclic and noncyclic photosynthetic pathway in which both ATP and NADPH could be formed from from a single input of electrons? A second limitation is that these bacterial systems require compounds such as reduced sulfur to act as electron donors to reduce the oxidized reaction centers, but they are not necessarily widely found compounds. What would happen if a chlorophyllox molecule would have a reduction potential higher (more positive) than that of the molecular O2/H2O reaction? Answer: a planetary game changer.
Oxygenic photophosphorylation
Generation of NADPH and ATP
The overall function of light-dependent reactions is to transfer solar energy into chemical compounds, largely the the molecules NADPH and ATP. This energy supports the light-independent reactions and fuels the assembly of sugar molecules. The light-dependent reactions are depicted in Figures 6 and 7. Protein complexes and pigment molecules work together to produce NADPH and ATP.
Note: possible discussion
Step back a little. Why is it a reasonable goal to want to make NADPH and ATP? In the discussion of glycolysis and the TCA cycle, the goal was to make ATP and NADH. What is the key difference? Perhaps how those molecules will be used? Something else?
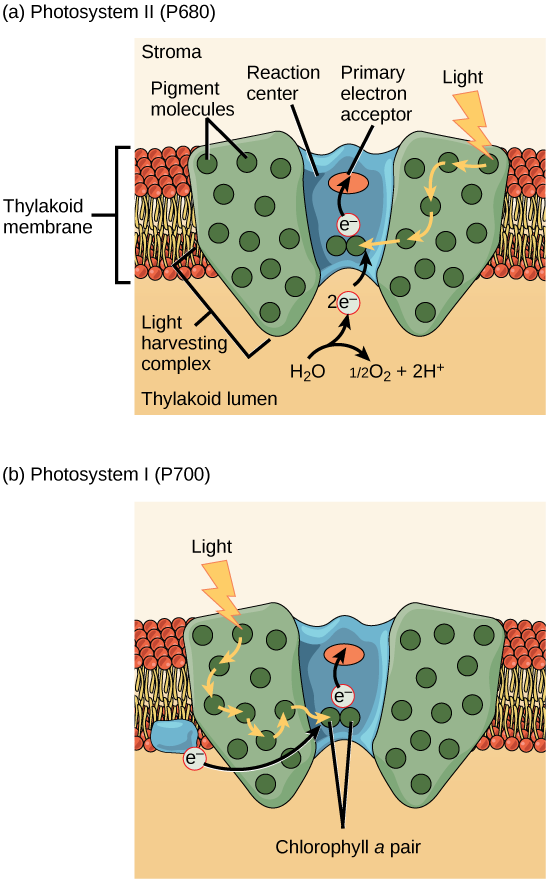
The actual step that transfers light energy into a biomolecule takes place in a multiprotein complex called a photosystem, two types of which are found embedded in the thylakoid membrane, photosystem II (PSII) and photosystem I (PSI). The two complexes differ on the basis of what they oxidize (that is, the source of the low energy electron supply) and what they reduce (the place to which they deliver their energized electrons).
Both photosystems have the same basic structure; a number of antenna proteins to which the chlorophyll molecules are bound surround the reaction center in which the photochemistry takes place. Each photosystem is serviced by the light-harvesting complex, which passes energy from sunlight to the reaction center; it consists of multiple antenna proteins that contain a mixture of 300–400 chlorophyll a and b molecules as well as other pigments like carotenoids. The absorption of a single photon—a distinct quantity or “packet” of light—by any of the chlorophylls pushes that molecule into an excited state. In short, the light energy has now been captured by biological molecules but is not yet stored in any useful form. The captured energy is transferred from chlorophyll to chlorophyll until, eventually (after about a millionth of a second), it is delivered to the reaction center. Up to this point, only energy has been transferred between molecules, not electrons.
The reaction center contains a pair of chlorophyll a molecules with a special property. Those two chlorophylls can undergo oxidation upon excitation; they can actually give up an electron in a process called a photoactivation. It is at this step in the reaction center, this step in photophosphorylation, that light energy is transferred into an excited electron. All of the subsequent steps involve getting that electron onto the energy carrier NADPH for delivery to the Calvin cycle where the electron can be deposited onto carbon for long term storage in the form of a carbohydrate.
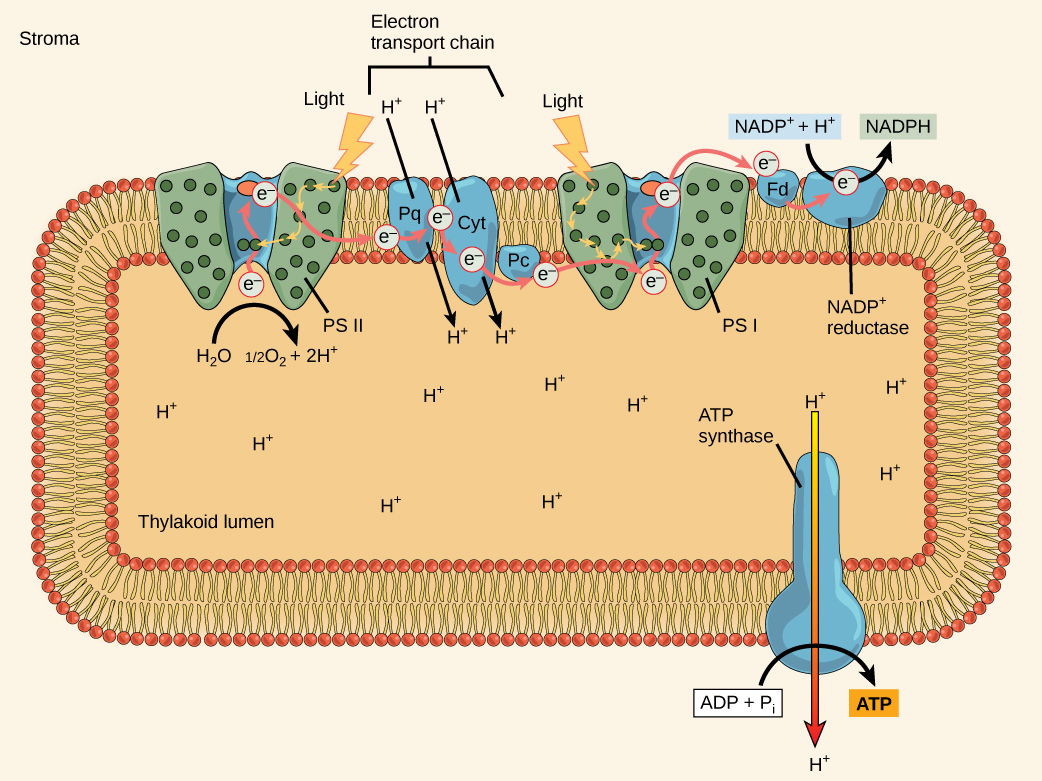
The Z-scheme
PSII and PSI are two major components of the photosynthetic electron transport chain, which also includes the cytochrome complex. The reaction center of PSII (called P680) delivers its high-energy electrons, one at a time, to a primary electron acceptor called pheophytin (Ph), and then sequentially to two bound plastoquinones QA and QB. Electrons are then transferred off of PSII onto a pool of mobile plastoquinones (Q pool) which then transfer the electrons to a protein complex called Cytochromeb6f. The cytochrome complex utilizes the red/ox transfers to pump proton across the thylakoyd membrane establishing a proton-motive force that can be used for the synthesis of ATP. Electrons leaving the Cytochrome are transferred to a copper-containing protein called plastocyanin (PC) which then transfers electrons to PSI (P700). P680’s missing electron is replaced by extracting an electron from water; thus, water is split and PSII is re-reduced after every photoactivation step. Just for the sake of sharing some numbers: Splitting one H2O molecule releases two electrons, two hydrogen atoms, and one atom of oxygen. Splitting two molecules of water is required to form one molecule of diatomic O2 gas. In plants, about ten percent of that oxygen is used by mitochondria in the leaf to support oxidative phosphorylation. The remainder escapes to the atmosphere where it is used by aerobic organisms to support respiration.
As electrons move through the proteins that reside between PSII and PSI, they take part in exergonic red/ox transfers. The free energy associated with the exergonic red/ox reaction is coupled to the endergonic transport of protons from the stromal side of the membrane to the thylakoid lumen by the cytochrome complex. Those hydrogen ions, plus the ones produced by splitting water, accumulate in the thylakoid lumen and create a proton motive force that will be used to drive the synthesis of ATP in a later step. Since the electrons on PSI now have a greater reduction potential than when they started their trek (it is important to note that unexcited PSI has a greater red/ox potential than NADP+/NADPH), they must be re-energized in PSI before getting deposited onto NADP+. Therefore, to complete this process, another photon must be absorbed by the PSI antenna. That energy is transferred to the PSI reaction center (called P700). P700 is then oxidized and sends an electron through several intermediate red/ox steps to NADP+ to form NADPH. Thus, PSII captures the energy in light and couples its transfer via red/ox reactions to the creation of a proton gradient. As already noted, the exergonic and controlled relaxation of this gradient can be coupled to the synthesis of ATP. PSI captures energy in light and couples that, through a series of red/ox reactions, to reduce NADP+ into NADPH. The two photosystems work in concert, in part, to guarantee that the production of NADPH will be in the right proportion to the production of ATP. Other mechanisms exist to fine tune that ratio to exactly match the chloroplast’s constantly changing energy needs.
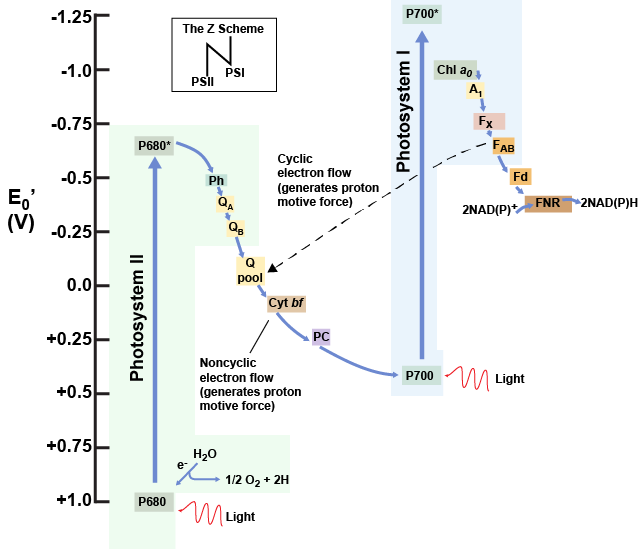
Figure 8. A diagram depicting the flow of electrons and the red/ox potentials of their carriers in oxygenic photosynthetic systems expressing both photosystem I (boxed in blue) and photosystem II (boxed in green). Ph = pheophytin; QA = bound plastoquinone, QB = more loosely associated plastoquinone; Q pool = mobile plastoquinone pool; Cyt bf = Cytochrome b6f complex; PC = plastocyanin; Chla0 = special chrolophyl; A1 = vitamin K; Fx and FAB = iron-sulfur centers; Fd = ferredoxin; FNR = ferredoxin-NADP reductase. Attribution: Marc T. Facciotti (own work)
Light Independent Reactions and Carbon Fixation
A short introduction
The general principle of carbon fixation is that some cells under certain conditions can take inorganic carbon, CO2 (also referred to as mineralized carbon), and reduce it to a usable cellular form. Most of us are aware that green plants can take up CO2 and produce O2 in a process known as photosynthesis. We have already discussed photophosphorylation, the ability of a cell to transfer light energy onto chemicals and ultimately to produce the energy carriers ATP and NADPH in a process known as the light reactions. In photosynthesis, the plant cells use the ATP and NADPH formed during photophosphorylation to reduce CO2 to sugar, (as we will see, specifically G3P) in what are called the dark reactions. While we appreciate that this process happens in green plants, photosynthesis had its evolutionary origins in the bacterial world. In this module we will go over the general reactions of the Calvin Cycle, a reductive pathway that incorporates CO2 into cellular material.
In photosynthetic bacteria, such as Cyanobacteria and purple non-sulfur bacteria, as well plants, the energy (ATP) and reducing power (NADPH) - a term used to describe electron carriers in their reduced state - obtained from photophosphorylation is coupled to "Carbon Fixation", the incorporation of inorganic carbon (CO2) into organic molecules; initially as glyceraldehyde-3-phosphate (G3P) and eventually into glucose. Organisms that can obtain all of their required carbon from an inorganic source (CO2) are referred to as autotrophs, while those organisms that require organic forms of carbon, such as glucose or amino acids, are referred to as heterotrophs. The biological pathway that leads to carbon fixation is called the Calvin Cycle and is a reductive pathway (consumes energy/uses electrons) which leads to the reduction of CO2 to G3P.
The Calvin Cycle: the reduction of CO2 to Glyceraldehyde 3-Phosphate
Figure 1. Light reactions harness energy from the sun to produce chemical bonds, ATP, and NADPH. These energy-carrying molecules are made in the stroma where carbon fixation takes place.
In plant cells, the Calvin cycle is located in the chloroplasts. While the process is similar in bacteria, there are no specific organelles that house the Calvin Cycle and the reactions occur in the cytoplasm around a complex membrane system derived from the plasma membrane. This intracellular membrane system can be quite complex and highly regulated. There is strong evidence that supports the hypothesis that the origin of chloroplasts from a symbiosis between cyanobacteria and early plant cells.
Stage 1: Carbon Fixation
In the stroma of plant chloroplasts, in addition to CO2, two other components are present to initiate the light-independent reactions: an enzyme called ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO), and three molecules of ribulose bisphosphate (RuBP), as shown in the figure below. Ribulose-1,5-bisphosphate (RuBP) is composed of five carbon atoms and includes two phosphates.
RuBisCO catalyzes a reaction between CO2 and RuBP. For each CO2 molecule that reacts with one RuBP, two molecules of another compound (3-PGA) form. PGA has three carbons and one phosphate. Each turn of the cycle involves only one RuBP and one carbon dioxide and forms two molecules of 3-PGA. The number of carbon atoms remains the same, as the atoms move to form new bonds during the reactions (3 atoms from 3CO2 + 15 atoms from 3RuBP = 18 atoms in 3 atoms of 3-PGA). This process is called carbon fixation, because CO2 is “fixed” from an inorganic form into an organic molecule.
Stage 2: Reduction
ATP and NADPH are used to convert the six molecules of 3-PGA into six molecules of a chemical called glyceraldehyde 3-phosphate (G3P) - a carbon compound that is also found in glycolysis. Six molecules of both ATP and NADPH are used in the process. The exergonic process of ATP hydrolysis is in effect driving the endergonic redox reactions, creating ADP and NADP+. Both of these "spent" molecules (ADP and NADP+) return to the nearby light-dependent reactions to be recycled back into ATP and NADPH.
Stage 3: Regeneration
Interestingly, at this point, only one of the G3P molecules leaves the Calvin cycle to contribute to the formation of other compounds needed by the organism. In plants, because the G3P exported from the Calvin cycle has three carbon atoms, it takes three “turns” of the Calvin cycle to fix enough net carbon to export one G3P. But each turn makes two G3Ps, thus three turns make six G3Ps. One is exported while the remaining five G3P molecules remain in the cycle and are used to regenerate RuBP, which enables the system to prepare for more CO2 to be fixed. Three more molecules of ATP are used in these regeneration reactions.