W2018_Bis2A_Lecture11_reading
- Page ID
- 25323
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Metabolism in BIS2A
Cellular metabolism represents roughly 1/3 of the BIS2A curriculum. While this may seem like a lot, we cover very little of what a classic course in metabolism (like BIS103) will cover, and an even smaller (really minuscule) fraction of the metabolism that occurs on the planet. What we do cover, however, is very important foundational knowledge. You will learn about some common chemical reactions that are associated with the transformation of life's molecular building blocks and about different core modes of energy transfer that you will encounter often in biology. The energy story and the design challenge rubrics introduced earlier will become increasingly important in these next few modules and beyond.
What have we learned? How will it relate to metabolism?
- We have focused on the identification and chemical properties of common biological functional groups. As we dive into metabolism, this will help you be familiar with and sometimes even predict the chemical nature/reactivity of compounds you have never seen before.
- We have practiced recognizing and classifying molecules into four major functional groups. This will help you as we begin to discuss how to build and break down these molecules.
- We have learned some basic thermodynamics, giving us a common set of concepts with which to discuss whether a biochemical reaction or process is likely to occur, and if so in which direction and how fast. This will be critical as we begin to consider some of the key types of reactions that take place in metabolism.
- We have learned and practiced the energy story rubric. This too will allow us to systematically examine new biochemical reactions and processes and to discuss them with a common language and approach that is consistent and reinforces the lessons we learned about thermodynamics.
A brief overview of this section
- You will be introduced to an important concept called reduction potential and you will be given the opportunity to use a redox tower. There is also a discussion on redox chemistry in your discussion manual. Make sure to use both resources.
- You will be introduced to two major players in metabolism, ATP and NADH. You will be expected to recognize their structures if shown on an exam.
- The metabolic pathway glycolysis will be covered in detail. Keep in mind that we want you to be able to look at any reaction and tell us an energy story of that reaction. By no means should you spend time trying to memorize these pathways (though it will help tremendously to remember some big picture things - these will be stressed). Often we will give you the pathway as a figure on the exams. Glycolysis ultimately produces 2 ATP via a process called substrate level phosphorylation, 2 NADH and 2 pyruvate compounds.
- We will use the reactions of the TCA cycle to create multiple examples of energy stories. The TCA cycle will also produce more ATP, NADH and completely oxidize glucose into CO2.
- We will look at an alternative pathway to that of the TCA cycle, fermentation. In fermentation for the first time we will see NADH used as a reactant in a metabolic reaction.
- We will follow NADH to the end of its journey, as it donates its electrons to the electron transport chain (ETC). In this module you will need to be able to use a redox tower. The ETC produces a proton gradient. No ATP is directly generated in this process. However, the proton gradient is then used by the cell (among other things) to run an enzyme called ATP synthase which catalyzes the reaction ADP + Pi --> ATP. This method of ATP production (called oxidative respiration) results in much more ATP being produced than substrate level phosphorylation.
- And finally, we will go through the process of photosynthesis.
Reduction-Oxidation Reactions
In this class, the majority of the oxidation/reduction reactions reactions that we discuss occur in the context of metabolic pathways (connected sets of metabolic reactions) where compounds may be consumed by the cell, broken down into smaller parts and then reassembled into larger macromolecules.
Lets start with some generic reactions
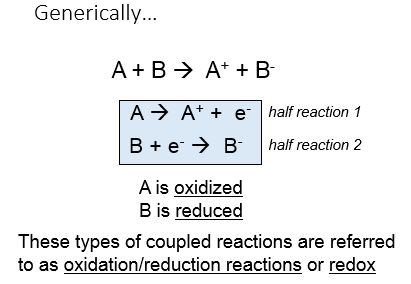
Transferring electrons between two compounds results in one of these compounds loosing an electron, and one of the compounds gaining an electron. For example, look at the figure below. If we use the energy story rubric to look at the overall reaction we can compare the before and after characteristics of the reactants and products. What happens to the matter (stuff) before and after the reaction? Compound A starts as neutral and becomes positively charged. Compound B starts as neutral and becomes negatively charged. Because electrons are negatively charged, we can follow the movement of electrons from compound A to B by looking at the change in charge. A looses an electron (becoming positively charged), and in so doing we say that A has become oxidized. Oxidation is associated with the loss of electron(s). B gains the electron (becoming negatively charged), and we say that B has become reduced. Reduction is associated with gain of electrons. We also know, since something happened that energy must have been either transferred and/or reorganized in this process and we'll consider this shortly.
Put another way, when an electron(s) is lost, or a molecule is oxidized, the electron(s) must then passed to another molecule. The molecule gaining the electron is said to be reduced. ***The oxidation and reduction reactions are always paired in what is known as an oxidation-reduction reaction (also called a red/ox reaction).****
In Bis2A we expect you to become familiar with this terminology. Try to learn it and learn to use it as soon as possible - we will use the terms frequently and will not have the time to define them each time.
Remember the Definitions:
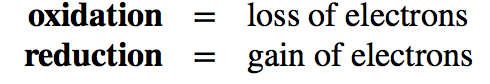
The Half Reaction
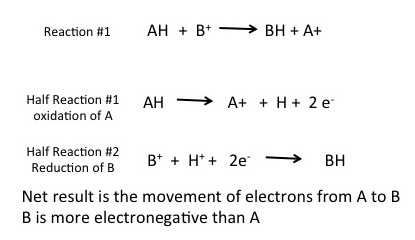
To formalize our common understanding of red/ox reactions, we introduce the concept of the half reaction. Two half reactions are required to make the full red/ox reaction. Each half reaction can be thought of as a description of what happens to one of the two molecules involved in the red/ox reaction. This is illustrated below. In this example compound AH is being oxidized by compound B+; electrons are moving from AH to B+ to generate A+ and BH. Each reaction can be thought of as two half reactions: Where AH is being oxidized and a second reaction where B+ is being reduced to BH. These two reactions are considered coupled, a term that indicates that these two reactions occur together, at the same time.
Possible discussion
If you consider a generic red/ox reaction and reflect back on the thermodynamic lectures what factor will determine whether a red/ox reaction will "go" in a particular direction spontaneously and what might determine its rate?
Reduction Potential
By convention we analyze and describe red/ox reactions with respect to reduction potentials, a term that quantitatively describes the "ability" of a compound to gain electrons. This value of the reduction potential is determined experimentally but for the purpose of this course we assume that the reader will accept that the reported values are reasonably correct. We can anthropomorphize the reduction potential by saying that it is related to the strength with which a compound can “attract” or “pull” or “capture” electrons. Not surprisingly this is is related to but not identical to electronegativity.
What is this intrinsic property to attract electrons?
Different compounds, based on their structure and atomic composition have intrinsic and distinct attractions for electrons. This quality is termed reduction potential or E0’and is a relative quantity (relative by comparison to some “standard” reaction). If a test compound has a stronger "attraction" to electrons than the standard (if the two competed the test compound would "take" electrons from the standard compound), we say that the test compound has a positive reduction potential whose magnitude is proportional to how much more it "wants" electrons than the standard compound. The relative strength of the compound in comparison to the standard is measured and reported in units of Volts (V)(sometimes written as electron volts or eV) or milliVolts (mV). The reference compound in most red/ox towers is H2.
Possible discussion
Rephrase for yourself: How do you describe or think about the difference between the concept of electronegativity and red/ox potential?
The Red/ox Tower
All kinds of compounds can participate in red/ox reactions. A tool has been developed to graphically tabulate red/ox half reactions based on their E0' values and to help us predict the direction of electron flow between potential electron donors and acceptors. Whether a particular compound can act as an electron donor (reductant) or electron acceptor (oxidant) depends critically on what other compound it is interacting with. The electron tower usually ranks a variety of common compounds (their half reactions) from most negative E0', compounds that readily get rid of electrons, to the most positive E0', compounds most likely to accept electrons. In addition, each half reaction is written by convention with the oxidized form on the left/followed by the reduced form on the right of the slash.
For example the half reaction for the reduction of NAD+ to NADH is written:
NAD+/NADH. In the tower below, the number of electrons that are transferred is also listed. For example the reduction of NAD+ to NADH involves two electrons, written in the table as 2e-.
An electron tower is shown below.
|
oxidized form |
reduced form |
n (electrons) |
Eo´ (volts) |
|
PS1* (ox) |
PS1* (red) |
- |
-1.20 |
|
Acetate + CO2 |
pyruvate |
2 |
-0.7 |
|
ferredoxin (ox) version 1 |
ferredoxin (red) version 1 |
1 |
-0.7 |
|
succinate + CO2 + 2H+ |
a-ketoglutarate + H2O |
2 |
-0.67 |
|
PSII* (ox) |
PSII* (red) |
- |
-0.67 |
|
P840* (ox) |
PS840* (red) |
- |
-0.67 |
|
acetate |
acetaldehyde |
2 |
-0.6 |
|
glycerate-3-P |
glyceraldehyde-3-P + H2O |
2 |
-0.55 |
|
O2 |
O2- |
1 |
-0.45 |
|
ferredoxin (ox) version 2 |
ferredoxin (red) version 2 |
1 |
-0.43 |
|
CO2 |
glucose |
24 |
-0.43 |
|
CO2 |
formate |
2 |
-0.42 |
|
2H+ |
H2 |
2 |
-0.42 |
|
α-ketoglutarate + CO2 + 2H+ |
isocitrate |
2 |
-0.38 |
|
acetoacetate |
b-hydroxybutyrate |
2 |
-0.35 |
|
Cystine |
cysteine |
2 |
-0.34 |
|
Pyruvate + CO2 |
malate |
2 |
-0.33 |
|
NAD+ + 2H+ |
NADH + H+ |
2 |
-0.32 |
|
NADP+ + 2H+ |
NADPH + H+ |
2 |
-0.32 |
|
Complex I FMN (enzyme bound) |
FMNH2 |
2 |
-0.3 |
|
Lipoic acid, (ox) |
Lipoic acid, (red) |
2 |
-0.29 |
|
1,3 bisphosphoglycerate + 2H+ |
glyceraldehyde-3-P + Pi |
2 |
-0.29 |
|
Glutathione, (ox) |
Glutathione, (red) |
2 |
-0.23 |
|
FAD+ (free) + 2H+ |
FADH2 |
2 |
-0.22 |
|
Acetaldehyde + 2H+ |
ethanol |
2 |
-0.2 |
|
Pyruvate + 2H+ |
lactate |
2 |
-0.19 |
|
Oxalacetate + 2H+ |
malate |
2 |
-0.17 |
|
α-ketoglutarate + NH4+ |
glutamate |
2 |
-0.14 |
|
FAD+ + 2H+ (bound) |
FADH2 (bound) |
2 |
0.003-0.09 |
|
Methylene blue, (ox) |
Methylene blue, (red) |
2 |
0.01 |
|
Fumarate + 2H+ |
succinate |
2 |
0.03 |
|
CoQ (Ubiquinone - UQ + H+) |
UQH. |
1 |
0.031 |
|
UQ + 2H+ |
UQH2 |
2 |
0.06 |
|
Dehydroascorbic acid |
ascorbic acid |
2 |
0.06 |
|
Plastoquinone; (ox) |
Plastoquinone; (red) |
- |
0.08 |
|
Ubiquinone; (ox) |
Ubiquinone; (red) |
2 |
0.1 |
|
Complex III Cytochrome b2; Fe3+ |
Cytochrome b2; Fe2+ |
1 |
0.12 |
|
Fe3+ (pH = 7) |
Fe2+ (pH = 7) |
1 |
0.20 |
|
Complex III Cytochrome c1; Fe3+ |
Cytochrome c1; Fe2+ |
1 |
0.22 |
|
Cytochrome c; Fe3+ |
Cytochrome c; Fe2+ |
1 |
0.25 |
|
Complex IV Cytochrome a; Fe3+ |
Cytochrome a; Fe2+ |
1 |
0.29 |
|
1/2 O2 + H2O |
H2O2 |
2 |
0.3 |
|
P840GS (ox) |
PS840GS (red) |
- |
0.33 |
|
Complex IV Cytochrome a3; Fe3+ |
Cytochrome a3; Fe2+ |
1 |
0.35 |
|
Ferricyanide |
ferrocyanide |
2 |
0.36 |
|
Cytochrome f; Fe3+ |
Cytochrome f; Fe2+ |
1 |
0.37 |
|
PSIGS (ox) |
PSIGS (red) |
. |
0.37 |
|
Nitrate |
nitrite |
1 |
0.42 |
|
Fe3+ (pH = 2) |
Fe2+ (pH = 2) |
1 |
0.77 |
|
1/2 O2 + 2H+ |
H2O |
2 |
0.816 |
|
PSIIGS (ox) |
PSIIGS (red) |
- |
1.10 |
|
* Excited State, after absorbing a photon of light GS Ground State, state prior to absorbing a photon of light PS1: Oxygenic photosystem I P840: Bacterial reaction center containing bacteriochlorophyll (anoxygenic) PSII: Oxygenic photosystem II |
|||
Video on electron tower
For a short video on how to use the electron tower in red/ox problems click here or below. This video was made by Dr. Easlon for Bis2A students. (This is quite informative.)
What is the relationship between ΔE0' and ΔG?
The question now becomes: how do we know if any given red/ox reaction is energetically spontaneous or not (exergonic or endergonic) and regardless of direction, what the free energy difference is? The answer lies in the difference in the reduction potentials of the two compounds. The difference in the reduction potential for the reaction or E0' for the reaction, is the difference between the E0' for the oxidant (the compound getting the electrons and causing the oxidation of the other compound) and the reductant (the compound losing the electrons). In our generic example below, AH is the reductant and B+ is the oxidant. Electrons are moving from AH to B+. Using the E0' of -0.32 for the reductant and +0.82 for the oxidant the total change in E0' or ΔE0' is 1.14 eV.
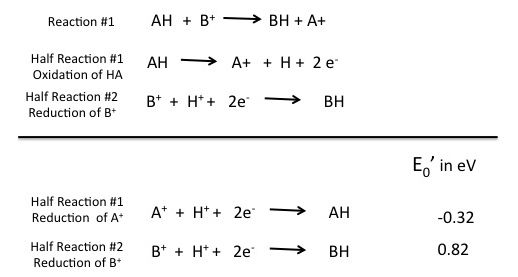
Figure 3. Generic red/ox reaction with half reactions written with reduction potential (E0') of the two half reactions indicated.
The change in ΔE0' correlates to changes in Gibbs free energy, ΔG. In general a large positive ΔE0' is proportional to a large negative ΔG. The reactions are exergonic and spontaneous. For a reaction to be exergonic the reaction needs to have a negative change in free energy or -ΔG, this will correspond to a positive ΔE0'. In other words, when electrons flow "downhill" in a red/ox reaction from a compound with a lower (more negative) reduction potential to a second compound with a larger (more positive) reduction potential, they release free energy. The greater the voltage, E0', between the two components, the greater the energy available when electron flow occurs. It is, in fact, possible to quantify the amount of free energy available. The relationship is given by the Nernst equation:
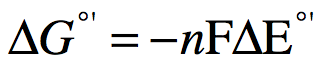
Figure 4. The Nernst equation relates free energy of a red/ox reaction to the difference in reduction potential between the reduced products of the reaction and oxidized reactant.
Attribution: Marc T. Facciotti
Where:
- n is the number of moles of electrons transferred
- F is the Faraday constant of 96.485 kJ/V. Sometimes it is given in units of kcal/V which is 23.062 kcal/V, which is the amount of energy (in kJ or kcal) released when one mole of electrons passes through a potential drop of 1 volt.
Note
What you should notice is that ΔG and ΔE have an inverse relationship: When ΔG is positive, ΔE is negative and when ΔG is negative, ΔE is positive. For additional review see the red/ox discussion in the Bis2A Discussion Manual.