SS1_2018_Lecture_06
( \newcommand{\kernel}{\mathrm{null}\,}\)
Oxidation of Pyruvate and the TCA Cycle
Overview of Pyruvate Metabolism and the TCA Cycle
Under appropriate conditions, pyruvate can be further oxidized. One of the most studied oxidation reactions involving pyruvate is a two-part reaction involving NAD+ and molecule called co-enzyme A, often abbreviated simply as "CoA". This reaction oxidizes pyruvate, leads to a loss of one carbon via decarboxylation, and creates a new molecule called acetyl-CoA. The resulting acetyl-CoA can enter several pathways for the biosynthesis of larger molecules or it can be routed to another pathway of central metabolism called the Citric Acid Cycle, sometimes also called the Krebs Cycle, or Tricarboxylic Acid (TCA) Cycle. Here the remaining two carbons in the acetyl group can either be further oxidized or serve again as precursors for the construction of various other molecules. We discuss these scenarios below.
The different fates of pyruvate and other end products of glycolysis
The glycolysis module left off with the end-products of glycolysis: 2 pyruvate molecules, 2 ATPs and 2 NADH molecules. This module and the module on fermentation explore what the cell can do with the pyruvate, ATP and NADH that were generated.
The fates of ATP and NADH
In general, ATP can be used for or coupled to a variety of cellular functions including biosynthesis, transport, replication etc. We will see many such examples throughout the course.
What to do with the NADH however, depends on the conditions under which the cell is growing. In some cases, the cell will opt to rapidly recycle NADH back into to NAD+. This occurs through a process called fermentation in which the electrons initially taken from the glucose derivatives are returned to more downstream products via another red/ox transfer (described in more detail in the module on fermentation). Alternatively, NADH can be recycled back into NAD+ by donating electrons to something known as an electron transport chain (this is covered in the module on respiration and electron transport).
The fate of cellular pyruvate
- Pyruvate can be used as a terminal electron acceptor (either directly or indirectly) in fermentation reactions, and is discussed in the fermentation module.
- Pyruvate could be secreted from the cell as a waste product.
- Pyruvate could be further oxidized to extract more free energy from this fuel.
- Pyruvate can serve as a valuable intermediate compound linking some of the core carbon processing metabolic pathways
The further oxidation of pyruvate
In respiring bacteria and archaea, the pyruvate is further oxidized in the cytoplasm. In aerobically respiring eukaryotic cells, the pyruvate molecules produced at the end of glycolysis are transported into mitochondria, which are sites of cellular respiration and house oxygen consuming electron transport chains (ETC in module on respiration and electron transport). Organisms from all three domains of life share similar mechanisms to further oxidize the pyruvate to CO2. First pyruvate is decarboxylated and covalently linked to co-enzyme A via a thioester linkage to form the molecule known as acetyl-CoA. While acetyl-CoA can feed into multiple other biochemical pathways we now consider its role in feeding the circular pathway known as the Tricarboxylic Acid Cycle, also referred to as the TCA cycle, the Citric Acid Cycle or the Krebs Cycle. This process is detailed below.
Conversion of Pyruvate into Acetyl-CoA
In a multi-step reaction catalyzed by the enzyme pyruvate dehydrogenase, pyruvate is oxidized by NAD+, decarboxylated, and covalently linked to a molecule of co-enzyme A via a thioester bond. The release of the carbon dioxide is important here, this reaction often results in a loss of mass from the cell as the CO2 will diffuse or be transported out of the cell and become a waste product. In addition, one molecule of NAD+ is reduced to NADH during this process per molecule of pyruvate oxidized. Remember: there are two pyruvate molecules produced at the end of glycolysis for every molecule of glucose metabolized; thus, if both of these pyruvate molecules are oxidized to acetyo-CoA two of the original six carbons will be converted to waste.
Suggested discussion
We have already discussed the formation of a thioester bond in another unit and lecture. Where was this specifically? What was the energetic significance of this bond? What are the similarities and differences between this example (formation of thioester with CoA) and the previous example of this chemistry?

Suggested discussion
Describe the flow and transfer of energy in this reaction using good vocabulary - (e.g. reduced, oxidized, red/ox, endergonic, exergonic, thioester, etc. etc.). You can peer edit - someone can start a description, another person can make it better, another person can improve it more etc. . .
In the presence of a suitable terminal electron acceptor, acetyl CoA delivers (exchanges a bond) its acetyl group to a four-carbon molecule, oxaloacetate, to form citrate (designated the first compound in the cycle). This cycle is called by different names: the citric acid cycle (for the first intermediate formed—citric acid, or citrate), the TCA cycle (since citric acid or citrate and isocitrate are tricarboxylic acids), and the Krebs cycle, after Hans Krebs, who first identified the steps in the pathway in the 1930s in pigeon flight muscles.
The Tricarboxcylic Acid (TCA) Cycle
In bacteria and archaea reactions in the TCA cycle typically happen in the cytosol. In eukaryotes, the TCA cycle takes place in the matrix of mitochondria. Almost all (but not all) of the enzymes of the TCA cycle are water soluble (not in the membrane), with the single exception of the enzyme succinate dehydrogenase, which is embedded in the inner membrane of the mitochondrion (in eukaryotes). Unlike glycolysis, the TCA cycle is a closed loop: the last part of the pathway regenerates the compound used in the first step. The eight steps of the cycle are a series of red/ox, dehydration, hydration, and decarboxylation reactions that produce two carbon dioxide molecules, one ATP, and reduced forms of NADH and FADH2.
Figure 2. In the TCA cycle, the acetyl group from acetyl CoA is attached to a four-carbon oxaloacetate molecule to form a six-carbon citrate molecule. Through a series of steps, citrate is oxidized, releasing two carbon dioxide molecules for each acetyl group fed into the cycle. In the process, three NAD+ molecules are reduced to NADH, one FAD+ molecule is reduced to FADH2, and one ATP or GTP (depending on the cell type) is produced (by substrate-level phosphorylation). Because the final product of the TCA cycle is also the first reactant, the cycle runs continuously in the presence of sufficient reactants.
Attribution: “Yikrazuul”/Wikimedia Commons (modified)
Note
We are explicitly making reference to eukaryotes, bacteria and archaea when we discuss the location of the TCA cycle because many beginning students of biology tend to exclusively associate the TCA cycle with mitochondria. Yes, the TCA cycle occurs in the mitochondria of eukaryotic cells. However, this pathway is not exclusive to eukaryotes; it occurs in bacteria and archaea too!
Steps in the TCA Cycle
Step 1:
The first step of the cycle is a condensation reaction involving the two-carbon acetyl group of acetyl-CoA with one four-carbon molecule of oxaloacetate. The products of this reaction are the six-carbon molecule citrate and free co-enzyme A. This step is considered irreversible because it is so highly exergonic. Moreover, the rate of this reaction is controlled through negative feedback by ATP. If ATP levels increase, the rate of this reaction decreases. If ATP is in short supply, the rate increases. If not already, the reason will become evident shortly.
Step 2:
In step two, citrate loses one water molecule and gains another as citrate is converted into its isomer, isocitrate.
Step 3:
In step three, isocitrate is oxidized by NAD+ and decarboxylated. Keep track of the carbons! This carbon now more than likely leaves the cell as waste and is no longer available for building new biomolecules. The oxidation of isocitrate therefore produces a five-carbon molecule, α-ketoglutarate, a molecule of CO2 and NADH. This step is also regulated by negative feedback from ATP and NADH, and via positive feedback from ADP.
Step 4:
Step 4 is catalyzed by the enzyme succinate dehydrogenase. Here, α-ketoglutarate is further oxidized by NAD+. This oxidation again leads to a decarboxylation and thus the loss of another carbon as waste. So far two carbons have come into the cycle from acetyl-CoA and two have left as CO2. At this stage, there is no net gain of carbons assimilated from the glucose molecules that are oxidized to this stage of metabolism. Unlike the previous step however succinate dehydrogenase - like pyruvate dehydrogenase before it - couples the free energy of the exergonic red/ox and decarboxylation reaction to drive the formation of a thioester bond between the substrate co-enzyme A and succinate (what is left after the decarboxylation). Succinate dehydrogenase is regulated by feedback inhibition of ATP, succinyl-CoA, and NADH.
Suggested discussion
We have seen several steps in this and other pathways that are regulated by allosteric feedback mechanisms. Is there something(s) in common about these steps in the TCA cycle? Why might these be good steps to regulate?
Suggested discussion
The thioester bond has reappeared! Use the terms we've been learning (e.g. reduction, oxidation, coupling, exergonic, endergonic etc.) to describe the formation of this bond and below its hydrolysis.
Step 5:
In step five, a substrate level phosphorylation event occurs. Here an inorganic phosphate (Pi) is added to GDP or ADP to form GTP (an ATP equivalent for our purposes) or ATP. The energy that drives this substrate level phosphorylation event comes from the hydrolysis of the CoA molecule from succinyl~CoA to form succinate. Why is either GTP or ATP produced? In animal cells there are two isoenzymes (different forms of an enzyme that carries out the same reaction), for this step, depending upon the type of animal tissue in which those cells are found. One isozyme is found in tissues that use large amounts of ATP, such as heart and skeletal muscle. This isozyme produces ATP. The second isozyme of the enzyme is found in tissues that have a large number of anabolic pathways, such as liver. This isozyme produces GTP. GTP is energetically equivalent to ATP; however, its use is more restricted. In particular, the process of protein synthesis primarily uses GTP. Most bacterial systems produce GTP in this reaction.
Step 6:
Step six is another red/ox reactions in which succinate is oxidized by FAD+ into fumarate. Two hydrogen atoms are transferred to FAD+, producing FADH2. The difference in reduction potential between the fumarate/succinate and NAD+/NADH half reactions is insufficient to make NAD+ a suitable reagent for oxidizing succinate with NAD+ under cellular conditions. However, the difference in reduction potential with the FAD+/FADH2 half reaction is adequate to oxidize succinate and reduce FAD+. Unlike NAD+, FAD+ remains attached to the enzyme and transfers electrons to the electron transport chain directly. This process is made possible by the localization of the enzyme catalyzing this step inside the inner membrane of the mitochondrion or plasma membrane (depending on whether the organism in question is eukaryotic or not).
Step 7:
Water is added to fumarate during step seven, and malate is produced. The last step in the citric acid cycle regenerates oxaloacetate by oxidizing malate with NAD+. Another molecule of NADH is produced in the process.
Summary
Note that this process (oxidation of pyruvate to Acetyl-CoA followed by one "turn" of the TCA cycle) completely oxidizes 1 molecule of pyruvate, a 3 carbon organic acid, to 3 molecules of CO2. Overall 4 molecules of NADH, 1 molecule of FADH2, and 1 molecule of GTP (or ATP) are also produced. For respiring organisms this is a significant mode of energy extraction, since each molecule of NADH and FAD2 can feed directly into the electron transport chain, and as we will soon see, the subsequent red/ox reactions that are driven by this process will indirectly power the synthesis of ATP. The discussion so far suggests that the TCA cycle is primarily an energy extracting pathway; evolved to extract or convert as much potential energy from organic molecules to a form that cells can use, ATP (or the equivalent) or an energized membrane. However, - and let us not forget - the other important outcome of evolving this pathway is the ability to produce several precursor or substrate molecules necessary for various catabolic reactions (this pathway provides some of the early building blocks to make bigger molecules). As we will discuss below, there is a strong link between carbon metabolism and energy metabolism.
Exercise
TCA Energy Stories
Work on building some energy stories yourself
There are a few interesting reactions that involve large transfers of energy and rearrangements of matter. Pick a few. Rewrite a reaction in your notes, and practice constructing an energy story. You now have the tools to discuss the energy redistribution in the context of broad ideas and terms like exergonic and endergonic. You also have the ability to begin discussing mechanism (how these reactions happen) by invoking enzyme catalysts. See your instructor and/or TA and check with you classmates to self-test on how you're doing.
Connections to Carbon Flow
One hypothesis that we have started exploring in this reading and in class is the idea that "central metabolism" evolved as a means of generating carbon precursors for catabolic reactions. Our hypothesis also states that as cells evolved, these reactions became linked into pathways: glycolysis and the TCA cycle, as a means to maximize their effectiveness for the cell. We can postulate that a side benefit to evolving this metabolic pathway was the generation of NADH from the complete oxidation of glucose - we saw the beginning of this idea when we discussed fermentation. We have already discussed how glycolysis not only provides ATP from substrate level phosphorylation, but also yields a net of 2 NADH molecules and 6 essential precursors: glucose-6-P, fructose-6-P, 3-phosphoglycerate, phosphoenolpyruvate, and of course, pyruvate. While ATP can be used by the cell directly as an energy source, NADH posses a problem and must be recycled back into NAD+, to keep the pathway in balance. As we see in detail in the fermentation module, the most ancient way cells deal with this problem is to use fermentation reactions to regenerate NAD+.
During the process of pyruvate oxidation via the TCA cycle 4 additional essential precursors are formed: acetyl~CoA, α-ketoglutarate, oxaloacetate, and succinyl~CoA. Three molecules of CO2 are lost and this represents a net loss of mass for the cell. These precursors, however, are substrates for a variety of catabolic reactions including the production of amino acids, fatty acids, and various co-factors, such as heme. This means that the rate of reactions through the TCA cycle will be sensitive to the concentrations of each metabolic intermediate (more on the thermodynamics in class). A metabolic intermediate is a compound that is produced by one reaction (a product) and then acts as a substrate for the next reaction. This also means that metabolic intermediates, in particular the 4 essential precursors, can be removed at any time for catabolic reactions, if there is a demand, changing the thermodynamics of the cycle.
Not all cells have a functional TCA cycle
Since all cells require the ability of make these precursor molecules, one might expect that all organisms would have a fully functional TCA cycle. In fact, the cells of many organisms DO NOT have all of the enzymes required to form a complete cycle - all cells, however, DO have the capability of making the 4 TCA cycle precursors noted in the previous paragraph. How can the cells make precursors and not have a full cycle? Remember that most of these reactions are freely reversible, so, if NAD+ is required to for the oxidation of pyruvate or acetyl~CoA, then the reverse reactions would require NADH. This process is often referred to as the reductive TCA cycle. To drive these reactions in reverse (with respect to the direction discussed above) requires energy, in this case carried by ATP and NADH. If you get ATP and NADH driving a pathway one direction, it stands to reason that driving it in reverse will require ATP and NADH as "inputs". So, organisms that do not have a full cycle can still make the 4 key metabolic precursors by using previously extracted energy and electrons (ATP and NADH) to drive some key steps in reverse.
Suggested discussion
Why might some organisms not have evolved a fully oxidative TCA cycle? Remember, cells need to keep a balance in the NAD+ to NADH ratio as well as the [ATP]/[AMP]/[ADP] ratios.
Additional Links
Here are some additional links to videos and pages that you may find useful.
Chemwiki Links
- Chemwiki TCA cycle - link down until key content corrections are made to the resource
Introduction to Respiration and Electron Transport Chains
General Overview and Points to Keep In Mind
In the next few modules, we start to learn about the process of respiration and the roles that electron transport chains play in this process. A definition of the word "respiration" that most people are familiar with is "the act of breathing". When we breath, air including molecular oxygen is brought into our lungs from outside of the body, the oxygen then becomes reduced, and waste products, including the reduced oxygen in the form of water, are exhaled. More generically, some reactant comes into the organism and then gets reduced and leaves the body as a waste product.
This generic idea, in a nutshell, can be generally applied across biology. Note that oxygen need not always be the compound that brought in, reduced, and dumped as waste. The compounds onto which the electrons that are "dumped" are more specifically known as "terminal electron acceptors." The molecules from which the electrons originate vary greatly across biology (we have only looked at one possible source - the reduced carbon-based molecule glucose).
In between the original electron source and the terminal electron acceptor are a series of biochemical reactions involving at least one red/ox reaction. These red/ox reactions harvest energy for the cell by coupling exergonic red/ox reaction to an energy-requiring reaction in the cell. In respiration, a special set of enzymes carry out a linked series of red/ox reactions that ultimately transfer electrons to the terminal electron acceptor.
These "chains" of red/ox enzymes and electron carriers are called electron transport chains (ETC). In aerobically respiring eukaryotic cells the ETC is composed of four large, multi-protein complexes embedded in the inner mitochondrial membrane and two small diffusible electron carriers shuttling electrons between them. The electrons are passed from enzyme to enzyme through a series of red/ox reactions. These reactions couple exergonic red/ox reactions to the endergonic transport of hydrogen ions across the inner mitochondrial membrane. This process contributes to the creation of a transmembrane electrochemical gradient. The electrons passing through the ETC gradually lose potential energy up until the point they are deposited on the terminal electron acceptor which is typically removed as waste from the cell. When oxygen acts as the final electron acceptor, the free energy difference of this multi-step red/ox process is ~ -60 kcal/mol when NADH donates electrons or ~ -45 kcal/mol when FADH2 donates.
Note: Oxygen is not the only, nor most frequently used, terminal electron acceptor in nature
Recall, that we use oxygen as an example of only one of numerous possible terminal electron acceptors that can be found in nature. The free energy differences associated with respiration in anaerobic organisms will be different.
In prior modules we discussed the general concept of red/ox reactions in biology and introduced the Electron Tower, a tool to help you understand red/ox chemistry and to estimate the direction and magnitude of potential energy differences for various red/ox couples. In later modules, substrate level phosphorylation and fermentation were discussed and we saw how exergonic red/ox reactions could be directly coupled by enzymes to the endergonic synthesis of ATP.
These processes are hypothesized to be one of the oldest forms of energy production used by cells. In this section we discuss the next evolutionary advancement in cellular energy metabolism, oxidative phosphorylation. First and foremost recall that, oxidative phosphorylation does not imply the use of oxygen. Rather the term oxidative phosphorylation is used because this process of ATP synthesis relies on red/ox reactions to generate a electrochemical transmembrane potential that can then be used by the cell to do the work of ATP synthesis.
A Quick Overview of Principles Relevant to Electron Transport Chains
An ETC begins with the addition of electrons, donated from NADH, FADH2 or other reduced compounds. These electrons move through a series of electron transporters, enzymes that are embedded in a membrane, or other carriers that undergo red/ox reactions. The free energy transferred from these exergonic red/ox reactions is often coupled to the endergonic movement of protons across a membrane. Since the membrane is an effective barrier to charged species, this pumping results in an unequal accumulation of protons on either side of the membrane. This in turn "polarizes" or "charges" the membrane, with a net positive (protons) on one side of the membrane and a negative charge on the other side of the membrane. The separation of charge creates an electrical potential. In addition, the accumulation of protons also causes a pH gradient known as a chemical potential across the membrane. Together these two gradients (electrical and chemical) are called an electro-chemical gradient.
Review: The Electron Tower
Since red/ox chemistry is so central to the topic we begin with a quick review of the table of reduction potential - sometimes called the "red/ox tower" or "electron tower". You may hear your instructors use these terms interchangeably. As we discussed in previous modules, all kinds of compounds can participate in biological red/ox reactions. Making sense of all of this information and ranking potential red/ox pairs can be confusing. A tool has been developed to rate red/ox half reactions based on their reduction potentials or E0' values. Whether a particular compound can act as an electron donor (reductant) or electron acceptor (oxidant) depends on what other compound it is interacting with. The red/ox tower ranks a variety of common compounds (their half reactions) from most negative E0', compounds that readily get rid of electrons, to the most positive E0', compounds most likely to accept electrons. The tower organizes these half reactions based on the ability of electrons to accept electrons. In addition, in many red/ox towers each half reaction is written by convention with the oxidized form on the left followed by the reduced form to its right. The two forms may be either separated by a slash, for example the half reaction for the reduction of NAD+ to NADH is written: NAD+/NADH + 2e-, or by separate columns. An electron tower is shown below.
Note
Use the red/ox tower above as a reference guide to orient you as to the reduction potential of the various compounds in the ETC. Red/ox reactions may be either exergonic or endergonic depending on the relative red/ox potentials of the donor and acceptor. Also remember there are many different ways of looking at this conceptually; this type of red/ox tower is just one way.
Note: Language shortcuts reappear
In the red/ox table above some entries seem to be written in unconventional ways. For instance Cytochrome cox/red. There only appears to be one form listed. Why? This is another example of language shortcuts (likely because someone was too lazy to write cytochrome twice) that can be confusing - particularly to students. The notation above could be rewritten as Cytochrome cox/Cytochrome cred to indicate that the cytochrome c protein can exist in either and oxidized state Cytochrome cox or reduced state Cytochrome cred.
Review Red/ox Tower Video
For a short video on how to use the red/ox tower in red/ox problems click here. This video was made by Dr. Easlon for Bis2A students.
Using the red/ox tower: A tool to help understand electron transport chains
By convention the tower half reactions are written with the oxidized form of the compound on the left and the reduced form on the right. Notice that compounds such as glucose and hydrogen gas are excellent electron donors and have very low reduction potentials E0'. Compounds, such as oxygen and nitrite, whose half reactions have relatively high positive reduction potentials (E0') generally make good electron acceptors are found at the opposite end of the table.
Example: Menaquinone
Let's look at menaquinoneox/red. This compound sits in the middle of the red/ox tower with an half-reaction E0' value of -0.074 eV. Menaquinoneox can spontaneously (ΔG<0) accept electrons from reduced forms of compounds with lower half-reaction E0'. Such transfers form menaquinonered and the oxidized form of the original electron donor. In the table above, examples of compounds that could act as electron donors to menaquinone include FADH2, an E0' value of -0.22, or NADH, with an E0' value of -0.32 eV. Remember the reduced forms are on the right hand side of the red/ox pair.
Once menaquinone has been reduced, it can now spontaneously (ΔG<0) donate electrons to any compound with a higher half-reaction E0' value. Possible electron acceptors include cytochrome box with an E0' value of 0.035 eV; or ubiquinoneox with an E0' of 0.11 eV. Remember that the oxidized forms lie on the left side of the half reaction.
Electron Transport Chains
An electron transport chain, or ETC, is composed of a group of protein complexes in and around a membrane that help energetically couple a series of exergonic/spontaneous red/ox reactions to the endergonic pumping of protons across the membrane to generate an electrochemical gradient. This electrochemical gradient creates a free energy potential that is termed a proton motive force whose energetically "downhill" exergonic flow can later be coupled to a variety of cellular processes.
ETC overview
Step 1: Electrons enter the ETC from an electron donor, such as NADH or FADH2, which are generated during a variety of catabolic reactions, including those associated glucose oxidation. Depending on the number and types of electron carriers of the ETC being used by an organism, electrons can enter at a variety of places in the electron transport chain. Entry of electrons at a specific "spot" in the ETC depends upon the respective reduction potentials of the electron donors and acceptors.
Step 2: After the first red/ox reaction, the initial electron donor will become oxidized and the electron acceptor will become reduced. The difference in red/ox potential between the electron acceptor and donor is related to ΔG by the relationship ΔG = -nFΔE, where n = the number of electrons transferred and F = Faraday's constant. The larger a positive ΔE, the more exergonic the red/ox reaction is.
Step 3: If sufficient energy is transferred during an exergonic red/ox step, the electron carrier may couple this negative change in free energy to the endergonic process of transporting a proton from one side of the membrane to the other.
Step 4: After usually multiple red/ox transfers, the electron is delivered to a molecule known as the terminal electron acceptor. In the case of humans, the terminal electron acceptor is oxygen. However, there are many, many, many, other possible electron acceptors in nature; see below.
Note: possible discussion
Electrons entering the ETC do not have to come from NADH or FADH2. Many other compounds can serve as electron donors; the only requirements are (1) that there exists an enzyme that can oxidize the electron donor and then reduce another compound, and (2) that the ∆E0' is positive (e.g., ΔG<0). Even a small amounts of free energy transfers can add up. For example, there are bacteria that use H2 as an electron donor. This is not too difficult to believe because the half reaction 2H+ + 2 e-/H2 has a reduction potential (E0') of -0.42 V. If these electrons are eventually delivered to oxygen, then the ΔE0' of the reaction is 1.24 V, which corresponds to a large negative ΔG (-ΔG). Alternatively, there are some bacteria that can oxidize iron, Fe2+ at pH 7 to Fe3+ with a reduction potential (E0') of + 0.2 V. These bacteria use oxygen as their terminal electron acceptor, and, in this case, the ΔE0' of the reaction is approximately 0.62 V. This still produces a -ΔG. The bottom line is that, depending on the electron donor and acceptor that the organism uses, a little or a lot of energy can be transferred and used by the cell per electrons donated to the electron transport chain.
What are the complexes of the ETC?
ETCs are made up of a series (at least one) of membrane-associated red/ox proteins or (some are integral) protein complexes (complex = more than one protein arranged in a quaternary structure) that move electrons from a donor source, such as NADH, to a final terminal electron acceptor, such as oxygen. This particular donor/terminal acceptor pair is the primary one used in human mitochondria. Each electron transfer in the ETC requires a reduced substrate as an electron donor and an oxidized substrate as the electron acceptor. In most cases, the electron acceptor is a member of the enzyme complex itsef. Once the complex is reduced, the complex can serve as an electron donor for the next reaction.
How do ETC complexes transfer electrons?
As previously mentioned, the ETC is composed of a series of protein complexes that undergo a series of linked red/ox reactions. These complexes are in fact multi-protein enzyme complexes referred to as oxidoreductases or simply, reductases. The one exception to this naming convention is the terminal complex in aerobic respiration that uses molecular oxygen as the terminal electron acceptor. That enzyme complex is referred to as an oxidase. Red/ox reactions in these complexes are typically carried out by a non-protein moiety called a prosthetic group. The prosthetic groups are directly involved in the red/ox reactions being catalyzed by their associated oxidoreductases. In general, these prosthetic groups can be divided into two general types: those that carry both electrons and protons and those that only carry electrons.
Note
This use of prosthetic groups by members of ETC is true for all of the electron carriers with the exception of quinones, which are a class of lipids that can directly be reduced or oxidized by the oxidoreductases. Both the Quinone(red) and the Quinone(ox) forms of these lipids are soluble within the membrane and can move from complex to complex to shuttle electrons.
The electron and proton carriers
- Flavoproteins (Fp), these proteins contain an organic prosthetic group called a flavin, which is the actual moiety that undergoes the oxidation/reduction reaction. FADH2 is an example of an Fp.
- Quinones are a family of lipids, which means they are soluble within the membrane.
- It should also be noted that NADH and NADPH are considered electron (2e-) and proton (2 H+) carriers.
Electron carriers
- Cytochromes are proteins that contain a heme prosthetic group. The heme is capable of carrying a single electron.
- Iron-Sulfur proteins contain a nonheme iron-sulfur cluster that can carry an electron. The prosthetic group is often abbreviated as Fe-S
Aerobic versus anaerobic respiration
We humans use oxygen as the terminal electron acceptor for the ETCs in our cells. This is also the case for many of the organisms we intentionally and frequently interact with (e.g. our classmates, pets, food animals, etc). We breath in oxygen; our cells take it up and transport it into the mitochondria where it is used as the final acceptor of electrons from our electron transport chains. That process - because oxygen is used as the terminal electron acceptor - is called aerobic respiration.
While we may use oxygen as the terminal electron acceptor for our respiratory chains, this is not the only mode of respiration on the planet. Indeed, the more general processes of respiration evolved at a time when oxygen was not a major component of the atmosphere. As a consequence, many organisms can use a variety of compounds including nitrate (NO3-), nitrite (NO2-), even iron (Fe3+) as terminal electron acceptors. When oxygen is NOT the terminal electron acceptor, the process is referred to as anaerobic respiration. Therefore, respiration or oxidative phosphorylation does not require oxygen at all; it simply requires a compound with a high enough reduction potential to act as a terminal electron acceptor, accepting electrons from one of the complexes within the ETC.
The ability of some organisms to vary their terminal electron acceptor provides metabolic flexibility and can ensure better survival if any given terminal acceptor is in limited supply. Think about this: in the absence of oxygen, we die; but other organisms can use a different terminal electron acceptor when conditions change in order to survive.
A generic example: A simple, two-complex ETC
The figure below depicts a generic electron transport chain, composed of two integral membrane complexes; Complex I(ox) and Complex II(ox). A reduced electron donor, designated DH (such as NADH or FADH2) reduces Complex I(ox), giving rise to the oxidized form D (such as NAD+ or FAD+). Simultaneously, a prosthetic group within Complex I is now reduced (accepts the electrons). In this example, the red/ox reaction is exergonic and the free energy difference is coupled by the enzymes in Complex I to the endergonic translocation of a proton from one side of the membrane to the other. The net result is that one surface of the membrane becomes more negatively charged, due to an excess of hydroxyl ions (OH-), and the other side becomes positively charged due to an increase in protons on the other side. Complex I(red) can now reduce a mobile electron carrier Q, which will then move through the membrane and transfer the electron(s) to the prosthetic group of Complex II(red). Electrons pass from Complex I to Q then from Q to Complex II via thermodynamically spontaneous red/ox reactions, regenerating Complex I(ox), which can repeat the previous process. Complex II(red) then reduces A, the terminal electron acceptor to regenerate Complex II(ox) and create the reduced form of the terminal electron acceptor, AH. In this specific example, Complex II can also translocate a proton during the process. If A is molecular oxygen, AH represents water and the process would be considered to be a model of an aerobic ETC. By contrast, if A is nitrate, NO3-, then AH represents NO2- (nitrite) and this would be an example of an anaerobic ETC.

Figure 1. Generic 2 complex electron transport chain. In the figure, DH is the electron donor (donor reduced), and D is the donor oxidized. A is the oxidized terminal electron acceptor, and AH is the final product, the reduced form of the acceptor. As DH is oxidized to D, protons are translocated across the membrane, leaving an excess of hydroxyl ions (negatively charged) on one side of the membrane and protons (positively charged) on the other side of the membrane. The same reaction occurs in Complex II as the terminal electron acceptor is reduced to AH.
Attribution: Marc T. Facciotti (original work)
Exercise 1
Thought question
Based on the figure above, use an electron tower to figure out the difference in the electrical potential if (a) DH is NADH and A is O2, and (b) DH is NADH and A is NO3-. Which pairs of electron donor and terminal electron acceptor (a) or (b) "extract" the greatest amount of free energy?
Detailed look at aerobic respiration
The eukaryotic mitochondria has evolved a very efficient ETC. There are four complexes composed of proteins, labeled I through IV depicted in the figure below. The aggregation of these four complexes, together with associated mobile, accessory electron carriers, is called also called an electron transport chain. This type of electron transport chain is present in multiple copies in the inner mitochondrial membrane of eukaryotes.
Figure 2. The electron transport chain is a series of electron transporters embedded in the inner mitochondrial membrane that shuttles electrons from NADH and FADH2 to molecular oxygen. In the process, protons are pumped from the mitochondrial matrix to the intermembrane space, and oxygen is reduced to form water.
Complex I
To start, two electrons are carried to the first protein complex aboard NADH. This complex, labeled I in Figure 2, includes flavin mononucleotide (FMN) and iron-sulfur (Fe-S)-containing proteins. FMN, which is derived from vitamin B2, also called riboflavin, is one of several prosthetic groups or cofactors in the electron transport chain. Prosthetic groups are organic or inorganic, nonpeptide molecules bound to a protein that facilitate its function; prosthetic groups include coenzymes, which are the prosthetic groups of enzymes. The enzyme in Complex I is also called NADH dehydrogenase and is a very large protein containing 45 individual polypeptide chains. Complex I can pump four hydrogen ions across the membrane from the matrix into the intermembrane space thereby helping to generate and maintain a hydrogen ion gradient between the two compartments separated by the inner mitochondrial membrane.
Q and Complex II
Complex II directly receives FADH2, which does not pass through Complex I. The compound connecting the first and second complexes to the third is ubiquinone (Q). The Q molecule is lipid soluble and freely moves through the hydrophobic core of the membrane. Once it is reduced, (QH2), ubiquinone delivers its electrons to the next complex in the electron transport chain. Q receives the electrons derived from NADH from Complex I and the electrons derived from FADH2 from Complex II, succinate dehydrogenase. Since these electrons bypass and thus do not energize the proton pump in the first complex, fewer ATP molecules are made from the FADH2 electrons. As we will see in the following section, the number of ATP molecules ultimately obtained is directly proportional to the number of protons pumped across the inner mitochondrial membrane.
Complex III
The third complex is composed of cytochrome b, another Fe-S protein, Rieske center (2Fe-2S center), and cytochrome c proteins; this complex is also called cytochrome oxidoreductase. Cytochrome proteins have a prosthetic group of heme. The heme molecule is similar to the heme in hemoglobin, but it carries electrons, not oxygen. As a result, the iron ion at its core is reduced and oxidized as it passes the electrons, fluctuating between different oxidation states: Fe2+ (reduced) and Fe3+ (oxidized). The heme molecules in the cytochromes have slightly different characteristics due to the effects of the different proteins binding them, giving slightly different characteristics to each complex. Complex III pumps protons through the membrane and passes its electrons to cytochrome c for transport to the fourth complex of proteins and enzymes (cytochrome c is the acceptor of electrons from Q; however, whereas Q carries pairs of electrons, cytochrome c can accept only one at a time).
Complex IV
The fourth complex is composed of cytochrome proteins c, a, and a3. This complex contains two heme groups (one in each of the two Cytochromes, a, and a3) and three copper ions (a pair of CuA and one CuB in Cytochrome a3). The cytochromes hold an oxygen molecule very tightly between the iron and copper ions until the oxygen is completely reduced. The reduced oxygen then picks up two hydrogen ions from the surrounding medium to make water (H2O). The removal of the hydrogen ions from the system contributes to the ion gradient used in the process of chemiosmosis.
Chemiosmosis
In chemiosmosis, the free energy from the series of red/ox reactions just described is used to pump protons across the membrane. The uneven distribution of H+ ions across the membrane establishes both concentration and electrical gradients (thus, an electrochemical gradient), owing to the proton's positive charge and their aggregation on one side of the membrane.
If the membrane were open to diffusion by protons, the ions would tend to diffuse back across into the matrix, driven by their electrochemical gradient. Ions, however, cannot diffuse through the nonpolar regions of phospholipid membranes without the aid of ion channels. Similarly, protons in the intermembrane space can only traverse the inner mitochondrial membrane through an integral membrane protein called ATP synthase (depicted below). This complex protein acts as a tiny generator, turned by transfer of energy mediated by protons moving down their electrochemical gradient. The movement of this molecular machine (enzyme) serves to lower the activation energy of reaction and couples the exergonic transfer of energy associated with the movement of protons down their electrochemical gradient to the endergonic addition of a phosphate to ADP, forming ATP.
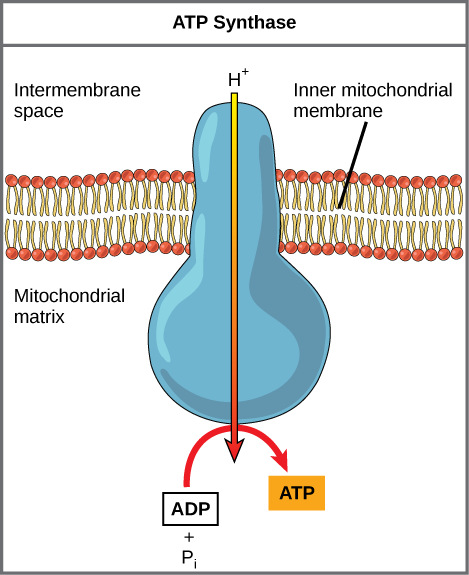
Figure 3. ATP synthase is a complex, molecular machine that uses a proton (H+) gradient to form ATP from ADP and inorganic phosphate (Pi).
Credit: modification of work by Klaus Hoffmeier
Note: possible discussion
Dinitrophenol (DNP) is a small chemical that serves to uncouple the flow of protons across the inner mitochondrial membrane to the ATP synthase, and thus the synthesis of ATP. DNP makes the membrane leaky to protons. It was used until 1938 as a weight-loss drug. What effect would you expect DNP to have on the difference in pH across both sides of the inner mitochondrial membrane? Why do you think this might be an effective weight-loss drug? Why might it be dangerous?
In healthy cells, chemiosmosis (depicted below) is used to generate 90 percent of the ATP made during aerobic glucose catabolism; it is also the method used in the light reactions of photosynthesis to harness the energy of sunlight in the process of photophosphorylation. Recall that the production of ATP using the process of chemiosmosis in mitochondria is called oxidative phosphorylation and that a similar process can occur in the membranes of bacterial and archaeal cells. The overall result of these reactions is the production of ATP from the energy of the electrons removed originally from a reduced organic molecule like glucose. In the aerobic example, these electrons ultimatel reduce oxygen and thereby create water.
Figure 4. In oxidative phosphorylation, the pH gradient formed by the electron transport chain is used by ATP synthase to form ATP in a Gram-bacteria.
Helpful link: How ATP is made from ATP synthase
Note: possible discussion
Cyanide inhibits cytochrome c oxidase, a component of the electron transport chain. If cyanide poisoning occurs, would you expect the pH of the intermembrane space to increase or decrease? What effect would cyanide have on ATP synthesis?
A Hypothesis for How ETC May Have Evolved
A proposed link between SLP/fermentation and the evolution of ETCs:
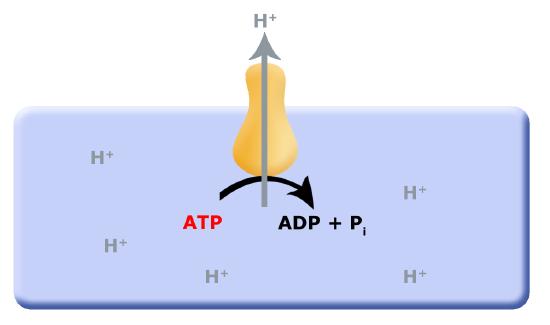
In a previous discussion of energy metabolism, we explored substrate level phosphorylation (SLP) and fermentation reactions. One of the questions in the discussion points for that discussion was: what are the short- and long-term consequences of SLP to the environment? We discussed how cells needed to co-evolve mechanisms in order to remove protons from the cytosol (interior of the cell), which led to the evolution of the F0F1-ATPase, a multi-subunit enzyme that translocates protons from the inside of the cell to the outside of the cell by hydrolyzing ATP, as shown below in the first picture below. This arrangement works as long as small reduced organic molecules are freely available, making SLP and fermentation advantageous. However, as these biological processes continue, the small reduced organic molecules begin to be used up, and their concentration decreases; this puts a demand on cells to be more efficient.
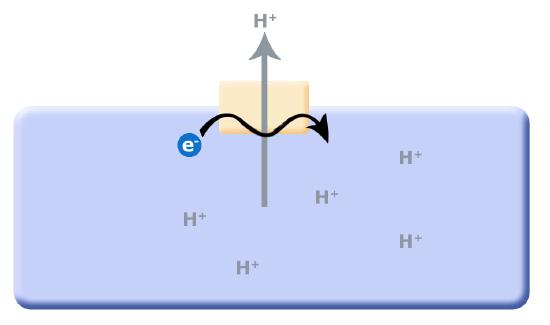
One source of potential "ATP waste" is in the removal of protons from the cell's cytosol; organisms that could find other mechanisms to expel accumulating protons while still preserving ATP could have a selective advantage. It is hypothesized that this selective evolutionary pressure potentially led to the evolution of the first membrane-bound proteins that used red/ox reactions as their energy source (depicted in second picture) to pump the accumulating protons. Enzymes and enzyme complexes with these properties exist today in the form of the electron transport complexes like Complex I, the NADH dehydrogenase.
Figure 1. Proposed evolution of an ATP dependent proton translocator
Figure 2. As small reduced organic molecules become limited, organisms that can find alternative mechanisms to remove protons from the cytosol may have an advantage. The evolution of a proton translocator that uses red/ox reactions rather than ATP hydrolysis could substitute for the ATPase.
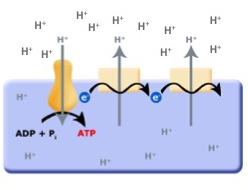
Continuing with this line of logic, if organisms evolved that could now use red/ox reactions to translocate protons across the membrane they would create a an electrochemical gradient, separating both charge (positive on the outside and negative on the inside; an electrical potential) and pH (low pH outside, higher pH inside). With excess protons on the outside of the cell membrane, and the F0F1-ATPase no longer consuming ATP to translocate protons, it is hypothesized that the electrochemical gradient could then be used to power the F0F1-ATPase "backwards" — that is, to form or produce ATP by using the energy in the charge/pH gradients set up by the red/ox pumps (as depicted below). This arrangement is called an electron transport chain (ETC).
Figure 3. The evolution of the ETC; the combination of the red/ox driven proton translocators coupled to the production of ATP by the F0F1-ATPase.
NoTE: Extended reading on the evolution of electron transport chains
If you're interested in the story of the evolution of electron transport chains, check out this more in-depth discussion of the topic at NCBI.