4.21: 2020_Spring_Bis2a_Facciotti_Lecture_21
- Page ID
- 27863
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)
Learning objectives associated with 2020_Spring_Bis2a_Facciotti_Lecture_21
|
Termination of replication
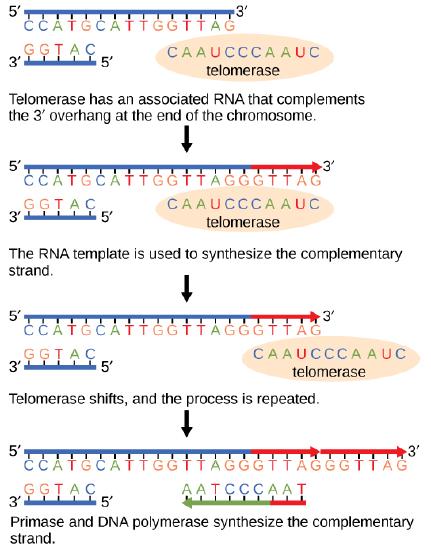
Telomeres and telomerase
The ends of replication in circular bacterial chromosomes pose few practical problems. However, the ends of linear eukaryotic chromosomes pose a specific problem for DNA replication. Because DNA polymerase can add nucleotides in only one direction (5' to 3'), the leading strand allows for continuous synthesis until the end of the chromosome
Figure 7. The ends of linear chromosomes
Telomerase is not active in adult somatic cells. Adult somatic cells that undergo cell division continue to have their telomeres shortened. This means that telomere shortening
Possible NB Discussion  Point
Point
Imagine that researchers have invented a drug for humans that upregulates telomerase activity. Would you be excited or would you be skeptical about being part of a clinical trial? Do you trust this drug? Can you think of any negative consequences to activating telomerase to a level that it would not “naturally” be at? Assuming scientists proved without doubt that their drug has no negative side effects -- what are some of the ethical questions you would consider?
Differences in DNA replication rates between bacteria and eukaryotes
DNA replication has
Table
| Differences between prokaryotic and eukaryotic replication | ||
|---|---|---|
| Property | Bacteria | Eukaryotes |
| Origin of replication | Single | Multiple |
| Rate of polymerization per polymerase | 1000 nucleotides/s | 50 to 100 nucleotides/s |
| Chromosome structure | Circular | Linear |
| Telomerase | Not present | |
Link to external resources
Click through a tutorial on DNA replication.
Replication design challenge: proofreading
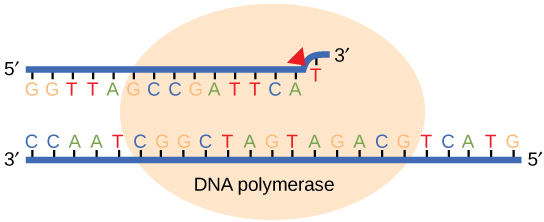
When the cell replicates its DNA, it does so in response to environmental signals that tell the cell it is time to divide. The ideal goal of DNA replication is to produce two identical copies of the double-stranded DNA template and to do it in an amount of time that does not pose an unduly high evolutionarily selective cost. This is a daunting task when you consider that there are ~6,500,000,000 base pairs in the human genome and ~4,500,000 base pairs in the genome of a typical E. coli strain and that Nature has determined that the cells must replicate within 24 hours and 20 minutes, respectively. In either case, many individual biochemical reactions need to take place.
While ideally replication would happen with perfect fidelity, DNA replication, like all other biochemical processes, is imperfect—bases may
If the polymerase detects that a newly added base has
We call errors that
Replication mistakes and DNA repair
Although DNA replication is typically a highly accurate process and proofreading DNA polymerases helps to keep the error rate low, mistakes still occur. Besides errors of replication, environmental damage may also occur to the DNA. Such uncorrected errors of replication or environmental DNA damage may lead to serious consequences. Therefore, Nature has evolved several mechanisms for repairing damaged or incorrectly synthesized DNA.
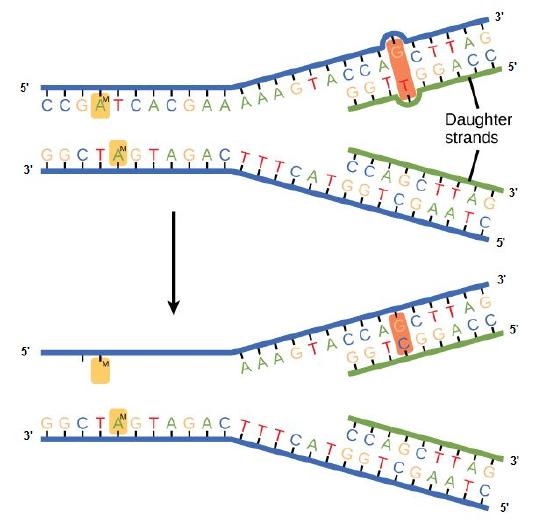
Mismatch repair
Errors not corrected during replication may instead get corrected after replication completes;
In
Figure 2. In mismatch repair, the incorrectly added base
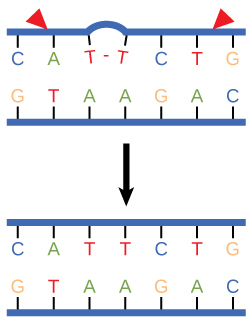
Nucleotide excision repair
Nucleotide excision repair enzymes replace incorrect bases by making a cut on both the 3' and 5' ends of the incorrect base. The entire segment of DNA
Figure 3. Nucleotide excision repairs thymine dimers. When exposed to UV, thymines lying
Consequences of errors in replication, transcription, and translation
Something key to think about:
Cells have evolved a variety of ways to make sure DNA errors are both detected and corrected. We have already discussed several of them. But why did so many mechanisms evolve? From proofreading by the various DNA-dependent DNA polymerases, to the complex repair systems. Such mechanisms did not evolve for errors in transcription or translation. If you are familiar with the processes of transcription and/or translation, think about what the consequences would be of an error in transcription. Would such an error affect the offspring? Would it be lethal to the cell? What about errors in translation? Ask the same questions about the process of translation. What would happen if