Sample Midterm 1
- Page ID
- 6204
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)\[\mathbf{BioSci\: 102,\\ Sample\: Study\: Problems\: for \:First\: Midterm}\]
1. What two factors determine how "good" (i. e. effective) a particular buffer solution is for maintaining a given pH?
a)
b)
2. The following question has two parts.
a) What is the final pH of a solution obtained by mixing 250 ml of 0.3 M acetic acid with 300 ml of 0.2 M KOH? (\(\mathrm{pK_{b}}\) of acetate = 9.24). (Show your work!)
b) What is the net charge on the acetic acid/acetate molecules at this pH?
3. Explain how you would make 2 liters of a 0.015 M buffer at pH 4.4 using crystalline disodium glutamate (MW = 191) and 0.9 M HCl. (use \(\mathrm{pK_{a}}\) values: \(\alpha\)-COOH = 2.2, side chain = 4.2, and \(\alpha\)-NH2 = 9.7)
4. A protein was completely digested with arg-c (an enzyme which cuts on the C-side of R residues) and an internal peptide (i. e. not derived from the N- or C- terminus) was purified. Determine its sequence from this information and the information given below.
a) Amino acid analysis of one mole of the peptide yields: 3 moles of cysteine, 1 mole of methionine, 3 moles of lysine, 1 mole of arginine and 1 mole of tryptophan.
b) Treatment of the intact peptide with trypsin gave rise to three different products; a dipeptide containing arg and trp, a dipeptide containing lys and cys, and a tripeptide containing cys, met and lys.
c) When the intact peptide was treated with Sanger's reagent (FDNB) and hydrolyzed with acid the derivatized products were DNP-cysteine and mono-DNP-lysine.
d) Treatment of the intact peptide with cyanogen bromide produced a pentapeptide containing cys, met, and lys; and a tetrapeptide containing arg, lys, trp and cys.
5. For each part of this question identify all amino acids being described (from among those normally occurring in proteins) by:
1) giving the one and three letter codes that are used to designate it (them) (note: each question may have more than one correct answer, please give them all) and;
2) drawing the full structure (of one correct answer) in the form that would predominate at pH 14.
a) An amino acid which is a structural isomer of another amino acid.
b) An amino acid which can absorb ultraviolet light and which has a side chain which can ionize at physiological pH.
c) An amino acid which is commonly the first amino acid in newly synthesized eukaryotic proteins.
d) The most basic of the 20 common amino acids (i. e. its side chain has the highest \(\mathrm{pK_{a}}\) value).
6. For the peptide shown below the numbers in parentheses indicate the \(\mathrm{pK_{a}}\) values for the adjacent ionizable groups. Answer the following three questions.
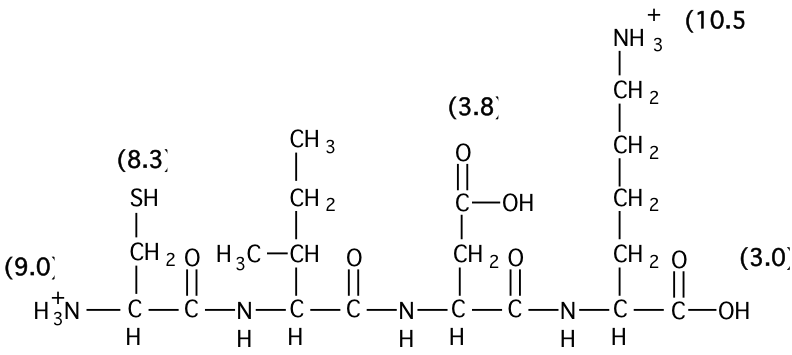
a) Calculate the charge of this peptide (to the nearest 0.1) at pH8.5.
b) Calculate the approximate pI of this peptide (assume that there are no interactions among the different ionizable groups).
c) Write out the name of this peptide using the one and three letter codes.
7. An enzyme catalyzed reaction was carried out in a solution buffered with 0.060 M histidine, pH 6.3. As a result of the reaction, the pH fell to 5.7. (use \(\mathrm{pK_{a}}\) values: \(\alpha\)-COOH = 1.82, imidazole side chain = 6.0, and \(\alpha\)-NH2 = 9.17).
a) How much acid (moles/liter) was produced as the result of the reaction?
b) What would the pH have been at the end of the reaction if it had been carried out in plain water instead of the 0.060 M histidine buffer?
8. Aldolase catalyzes the conversion of fructose-1,6-bisphosphate (F-1,6-BP) to dihydroxyacetone phosphate (DHAP) and glyceraldehyde-3-phosphate (G-3-P). The \(\mathrm{\Delta G^{\circ ‘}}\) for the reaction is +5.7 kcal/mol at \(\mathrm{37^{\circ C}}\).
a) The steady state concentrations given below were measured in red blood cells at \(\mathrm{37^{\circ C}}\). Calculate the \(\Delta G\) value for aldolase under these conditions.
\[\mathrm{[F-1,6-BP]=3.5\times 10^{-5}\: M}\]
\[\mathrm{[DHAP]=1.4\times 10^{-4}\: M}\]
\[\mathrm{[G-3-P]=2.1\times 10^{-5}\: M}\]
b) In which direction is the aldolase reaction proceeding under the steady state conditions given in a)? (Note: Your answer must be consistent with your answer in a)).
c) In which direction does the aldolase reaction proceed under biochemical standard state conditions?
d) During the reaction catalyzed by aldolase, the amino side chain of a lysine residue on the protein reacts with a keto group of F-1,6-BP. The lysine amino group must be unprotonated in order to react. Calculate the fraction of the lysine side chains that you would expect to be unprotonated at pH 7. (The \(\mathrm{pK_{a}}\)pKa of a lysine side chain is 10.5).

