4.4.1: Inheritance patterns for X-linked and Y-linked genes
- Page ID
- 34194
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)X-linked dominant inheritance
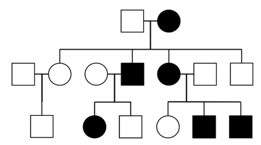
In X-linked dominant inheritance, the gene responsible for the disease is located on the X-chromosome, and the allele that causes the disease is dominant to the normal allele in XX individuals, who tend to be more frequently affected than XY individuals in the population because they have two copies of the X chromosome. However, not all pedigrees provide sufficient information to distinguish X-linked and autosomal dominance. One definitive indication that a trait is inherited on an autosome, and not the X chromosome, is that an affected XY sperm parent passes the disease to an XY child; this type of transmission is not possible with X-linked dominance, because XY individuals inherit their X chromosome from the XX egg parent.
X-linked recessive
Any XY individual that inherits an X-linked recessive disease allele will be affected by it (assuming complete penetrance), because they do not have a second copy of the X chromosome to provide a dominant allele. Therefore, in X-linked recessive inheritance, XY individuals, commonly males, tend to be affected more frequently than XX individuals, commonly females, in a population. In autosomal dominant and recessive traits, the sex of the individual does not change the probability of being affected, because all individuals have two copies of autosomes. Note, however, in the small sample sizes typical of human families, it is usually not possible to accurately determine whether one sex is affected more frequently than others. On the other hand, one feature of a pedigree that can be used to establish that an inheritance pattern is not X-linked recessive is the presence of an affected XX offspring from unaffected parents; because the sperm-parent would have a mutant X and therefore would also have been affected if the trait was X-linked recessive.

Figure \(\PageIndex{2}\): Some forms of colour blindness are inherited as X-linked recessive traits. Color blindness is diagnosed using tests such as this.
Ishihara Test. (Wikipedia-unknown-PD)
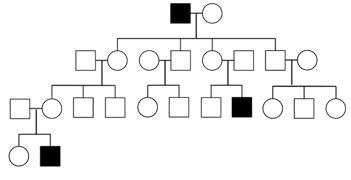
Y-linked Inheritance
Only individuals with a Y chromosome are affected in mammalian Y-linked inheritance and transmission can only occur from a sperm-producing parent. This is the easiest mode of inheritance to identify, but it is one of the rarest because there are so few genes located on the Y-chromosome. In 1960, the hairy-ear-rim phenotype seen in some Indian families was described as a Y-linked trait (Gates, RR https://science.sciencemag.org/content/132/3420/145.1.long) based on 20 pedigrees showing father to son transmission. In addition, the daughters of men with the hairy-ear-rim phenotype were unable to pass on the trait. However, questions remained about the accuracy of this conclusion and in 2004 a genotyping study found a lack of molecular evidence for Y linkage of this trait (Lee et al https://www.nature.com/articles/5201271.pdf).
Y-chromosome DNA polymorphisms can be used to follow the male lineage in large families or through ancient ancestral lineages. For example, the Y-chromosome of Mongolian ruler Genghis Khan (1162-1227 CE), and his male relatives, accounts for ~8% of the Y-chromosome lineage of men in Asia, or about 0.5% world wide (Zerjal et al, 2003).
Thinking about Y linkage
Aside from the size and small gene number on the Y chromosome, why are Y linked traits rare?
What functions do you think genes on the Y chromosome perform?
References
Lee AC, Kamalam A, Adams SM, Jobling MA. Molecular evidence for absence of Y-linkage of the Hairy Ears trait. Eur J Hum Genet. 2004 Dec;12(12):1077-9. doi: 10.1038/sj.ejhg.5201271. PMID: 15367914.
Zerjal T, Xue Y, Bertorelle G, Wells RS, Bao W, Zhu S, Qamar R, Ayub Q, Mohyuddin A, Fu S, Li P, Yuldasheva N, Ruzibakiev R, Xu J, Shu Q, Du R, Yang H, Hurles ME, Robinson E, Gerelsaikhan T, Dashnyam B, Mehdi SQ, Tyler-Smith C. The genetic legacy of the Mongols. Am J Hum Genet. 2003 Mar;72(3):717-21. doi: 10.1086/367774. Epub 2003 Jan 17. PMID: 12592608; PMCID: PMC1180246.

