3.4: Ames Test
- Page ID
- 63340
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Learning Objectives
- Explain the procedure of the Ames Test including use of histidine auxotrophs and replica plating.
Ames Test for Mutagenicity
The Ames test, developed by Bruce Ames (1928–) in the 1970s, is a method that uses bacteria for rapid, inexpensive screening of the carcinogenic potential of new chemical compounds. The test measures the mutation rate associated with exposure to the compound, which, if elevated, may indicate that exposure to this compound is associated with greater cancer risk. The Ames test uses as the test organism a strain of Salmonella typhimurium that is a histidine auxotroph, unable to synthesize its own histidine because of a mutation in an essential gene required for its synthesis. After exposure to a potential mutagen, these bacteria are plated onto a medium lacking histidine, and the number of mutants regaining the ability to synthesize histidine is recorded and compared with the number of such mutants that arise in the absence of the potential mutagen.
The Ames Test is possible because Ester Lederberg developed replica plating.
In replica plating, a piece of fabric is used to create an exact copy of bacterial colonies on two plates that differ in their media or environment. In the Ames test, the bacterial colonies can be grown in the presence and absence of histidine
Hear or read about how Ester Lederberg developed this critical technique in Genetics Unzipped (https://geneticsunzipped.com/news/2019/3/28/powder-puffs-and-plasmids-esther-lederberg).
Exercise \(\PageIndex{1}\)
Histidine auxotrophs cannot synthesize their own histidine, which is required for cell viability. Histidine mutants can be identified by replica plating and identifying colonies that can grow on plates with histidine but not on plates without histidine in the media. Which colonies in the figure below could have mutations in a histidine synthesis gene (his¯)?
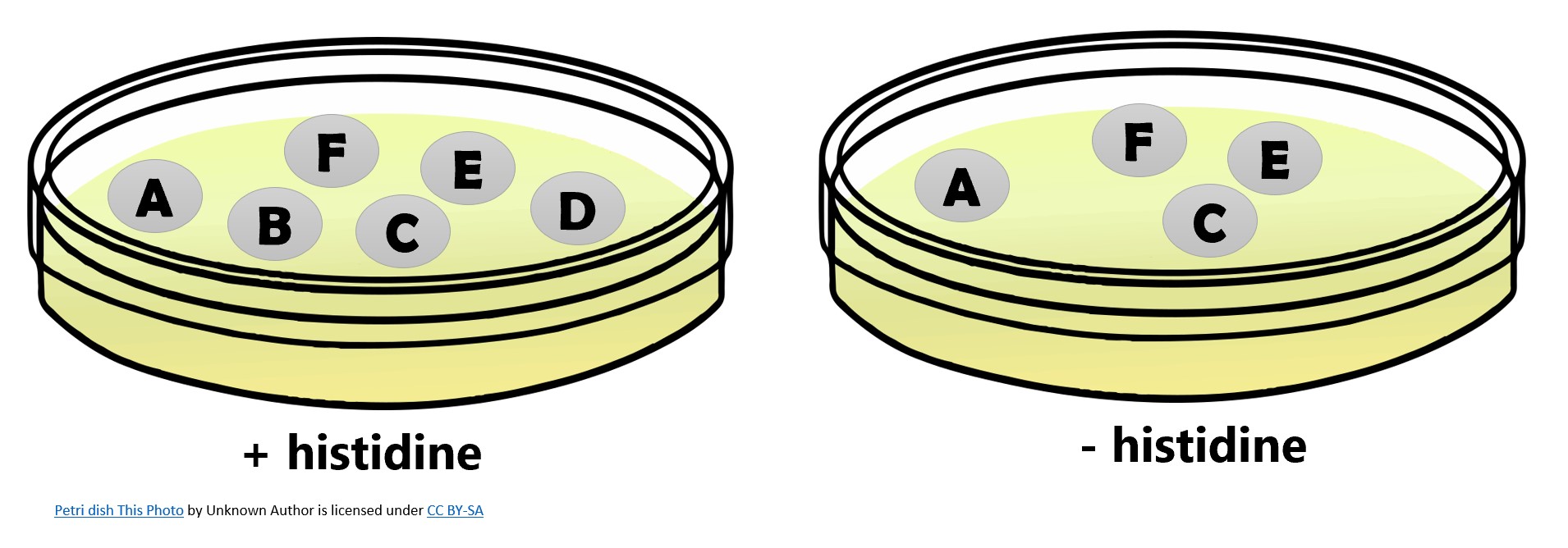
- Answer
-
The his¯ colonies are B and D.
Chemicals that are more mutagenic will bring about more mutants with restored histidine synthesis in the Ames test. Because many chemicals are not directly mutagenic but are metabolized to mutagenic forms by liver enzymes, rat liver extract is commonly included at the start of this experiment to mimic liver metabolism. After the Ames test is conducted, compounds identified as mutagenic are further tested for their potential carcinogenic properties by using other models, including animal models like mice and rats.
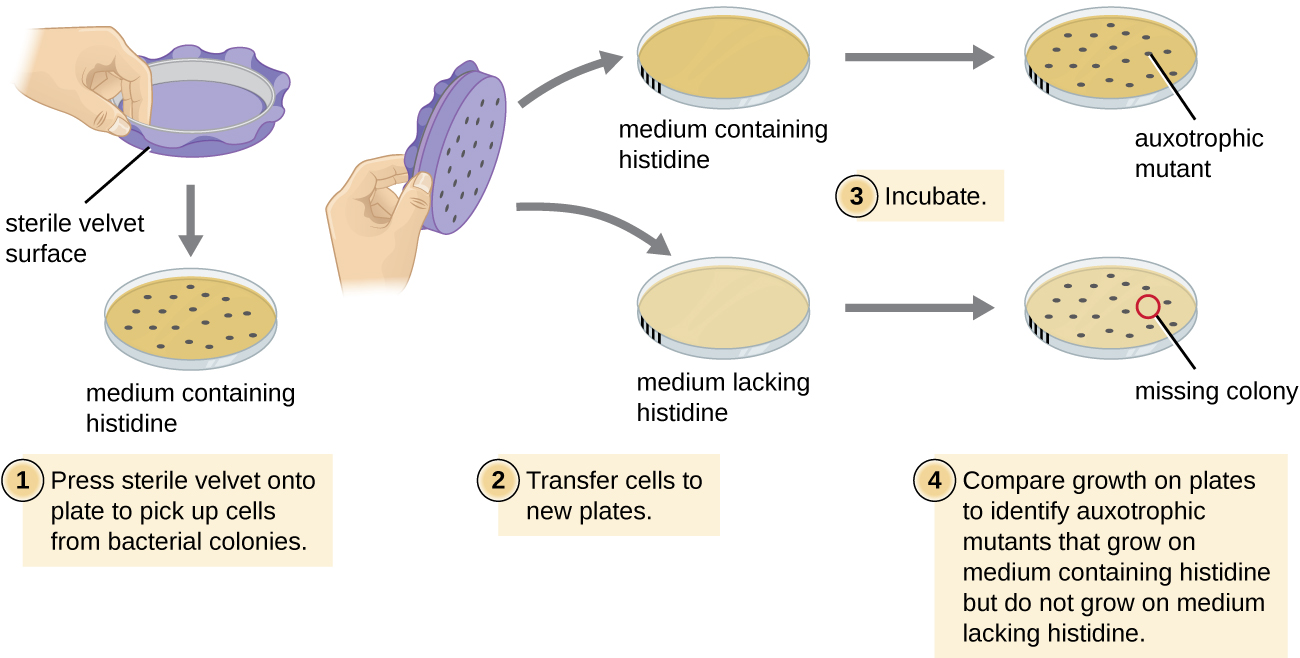
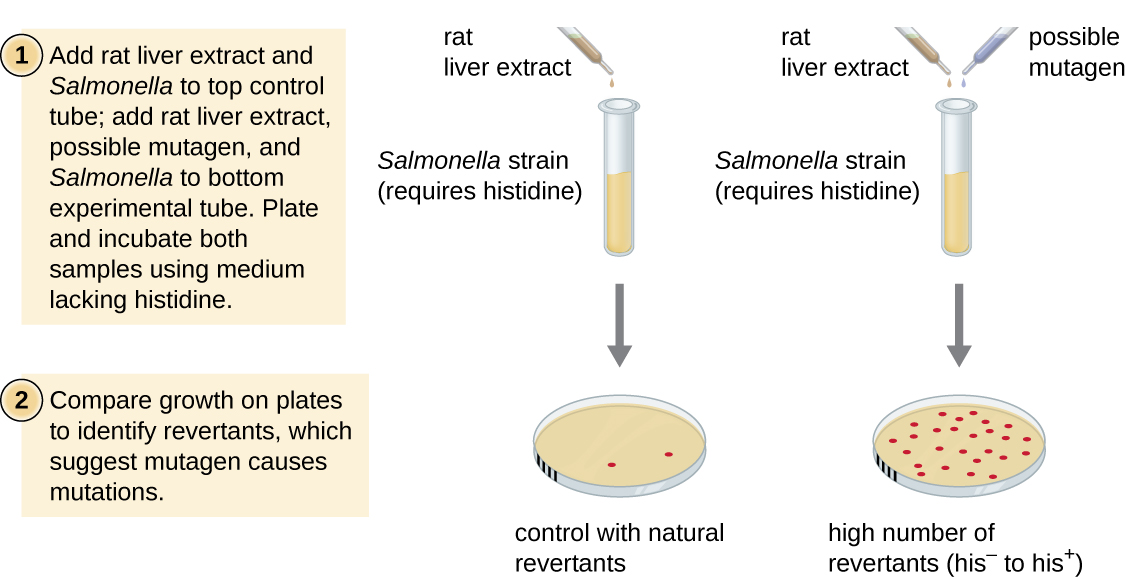
Figure \(\PageIndex{3}\): The Ames test is used to identify mutagenic, potentially carcinogenic chemicals. A Salmonella histidine auxotroph is used as the test strain, exposed to a potential mutagen or carcinogen. The number of reversion mutants capable of growing in the absence of supplied histidine is counted and compared with the number of natural reversion mutants that arise in the absence of the potential mutagen.
Questions \(\PageIndex{1}\)
- What mutation is used as an indicator of mutation rate in the Ames test?
- Why can the Ames test work as a test for carcinogenicity?
- Why do you think the Ames test is preferable to the use of animal models to screen chemical compounds for mutagenicity?
Exercise \(\PageIndex{2}\)
Which of the following regarding the Ames test is true?
A. It is used to identify newly formed auxotrophic mutants.
B. It is used to identify mutants with restored biosynthetic activity.
C. It is used to identify spontaneous mutants.
D. It is used to identify mutants lacking photoreactivation activity.
- Answer
-
B
Contributors and Attributions
Nina Parker, (Shenandoah University), Mark Schneegurt (Wichita State University), Anh-Hue Thi Tu (Georgia Southwestern State University), Philip Lister (Central New Mexico Community College), and Brian M. Forster (Saint Joseph’s University) with many contributing authors. Original content via Openstax (CC BY 4.0; Access for free at https://openstax.org/books/microbiology/pages/1-introduction)
Exercise \(\PageIndex{3}\)
Zinc oxide nanoparticles have been used in food processing and packaging as well as animal farming. Mittag and colleagues tested two different sizes of zinc oxide nanoparticles for mutagenicity using the Ames Test (2021). A small portion of their results is shown in the table.
- What is the genotype of the revertant colonies?
- Why do revertants occur in all conditions (even the negative control)?
- Does the data suggest that the ZnO nanoparticles are mutagenic?
- Does this data suggest that ZnO nanoparticles are not harmful to living things?
| Treatment | Average number of revertants/plate |
| Negative control (no nanoparticles) | 148 |
| 10 μg/plate ZnO NP (<50nm) | 148 |
| 10 μg/plate ZnO NP (<100nm) | 143 |
| Positive Control (a known mutagen) | 1136 |
Source: CC BY Mittag et al (2021) via https://www.mdpi.com/2305-6304/9/5/96/htm
- Answer
-
1. In an Ames Test, his¯ mutants are treated with the potential mutagen. Any DNA changes that restore the ability of the cell to synthesize histidine will produce his+ cells that have the ability to grow on media lacking histidine.
2. Random errors that occur during DNA replication are a constant source of new mutations; therefore the Ames Test is looking to detect a rate of mutations that occurs above this baseline level.
3. No. The number of revertants does not differ substantially from the negative control. (The complete paper reports that the p values from statistical tests of the data are not significant.)
4. Not necessarily. Other tests suggest that the nanoparticles could increase cell death. DNA damage is only one aspect to consider when determining whether a chemical is harmful to living things.
References
- Mittag A, Hoera C, Kämpfe A, Westermann M, Kuckelkorn J, Schneider T, Glei M. Cellular Uptake and Toxicological Effects of Differently Sized Zinc Oxide Nanoparticles in Intestinal Cells. Toxics. 2021 Apr 27;9(5):96. doi: 10.3390/toxics9050096. PMID: 33925422; PMCID: PMC8146923.

