6: Quantitative Determination of Serum
- Page ID
- 169818
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Introduction
A. Cholesterol has been implicated in the development of atherosclerosis and heart disease which are the leading causes of death in many countries. Since High-density lipoproteins (HDLs) and Low-density lipoproteins (LDLs) are the cholesterol-rich lipoprotein fractions, a high or low of cholesterol in the blood reflect a high or low level of these lipoproteins in our body. Measurement of blood cholesterol is important in the diagnosis of hyperlipoproteinmia and identification of the risk for cardiovascular disease.
B. Principle: The assay is based on the enzyme driven reaction that quantifies both cholesterol esters and free cholesterol. Cholesterol esters are hydrolyzed via cholesterol esterase into cholesterol, which is then oxidized by cholesterol oxidase into the ketone cholest-4-en-3-one plus hydrogen peroxide. The hydrogen peroxide is then detected with a highly specific colorimetric probe. Horseradish peroxidase catalyzes the reaction between the probe and hydrogen peroxide, which bind in a 1:1 ratio (Figure 1). Samples are compared to a known concentration of cholesterol standard in a 96-well microtiter plate format.

II. Materials and Methods:
A. Materials:
1. Assay Diluent (1X) prepared from Cell Biolab’s “Total Cholesterol Assay Kit”.
2.Cholesterol Standard: 250 μM cholesterol solution in 1X Assay diluent.
3. 50X Colorimetric Probe prepared from Cell Biolab’s “Total Cholesterol Assay Kit”.
4. Horseradish peroxidase (HRP).
5. Cholesterol Esterase
6. Cholesterol Oxidase
B. Equipment:
1.96-well microtiter plates
2. Spectrophotometric microplate reader capable of reading excitation in the 540-570 nm absorbance range.
C. Procedure:
1. Preparation of Cholesterol Standard Curve
Use this 250 μM solution to prepare a series of the cholesterol standards according to Table below.
|
Tube |
Final cholesterol Conc.(μM) |
250 uM cholesterol standard(μl) |
Assay diluent(μl) |
|
1 |
0 |
0 |
50 |
|
2 |
50 |
10 |
40 |
|
3 |
100 |
20 |
30 |
|
4 |
200 |
40 |
10 |
(配製標準樣品四管,如表0、50、100、200 uM,各一管 )
2. Preparation of Samples
Serum samples must be diluted at 1:50 (add 20μl of serum to 980μl of 1X Assay Diluent) and 1:100 (add 10μl of serum to 990μl of 1X Assay Diluent) with Assay Diluent. This will provide values within the range of the standard curve.
(準備未知濃度血清樣品,利用1X assay diluent分別將樣品稀釋50倍和100倍,各一管)
3. Preparation of the Reaction Reagents
|
Cholesterol oxidase |
16μl |
|
Horseradish peroxidase |
16μl |
|
Colorimetric Probe |
16μl |
|
Cholesterol esterase |
3.2μl |
|
1X Assay Diluent |
1000μl |
For best results, place the Reaction Reagent on ice and use within 30 minutes of preparation. Do not store the Cholesterol Reaction Reagent solution.
(依表格配製反應試劑。此試劑最好在30分鐘內使用完畢。
※請注意是二組共同配製一管共用,且配置前請將藥品先離心!
4. Add 50 μL of the diluted cholesterol standards or samples to a 96-well microtiter plate.
(將標準樣品和未知樣品分別取50μl加入96孔盤,四組共用一盤,如下圖)

5. Add 50 μL of the prepared Cholesterol Reaction Reagent to each well and mix the well contents thoroughly.
(在每個含有樣品的well加入50μl的反應試劑)
6. Cover the plate wells to protect the reaction from light. Incubate the plate for 45 minutes at 37ºC.(用鋁箔紙包覆96孔盤達避光效果,再將此96孔盤放到暖房或37℃烘箱45分鐘)
7. IMMEDIATELY read the plate with a spectrophotometric microplate reader in the 540 nm.
(45分鐘後立即使用微量光譜儀讀取波長540nm的吸光值)
8. Plot the Cholesterol Standard Curve.
(畫出標準曲線,並算出回歸曲線公式)
9. Calculate the concentration of cholesterol within samples by comparing the sample absorbance values to the cholesterol standard curve.
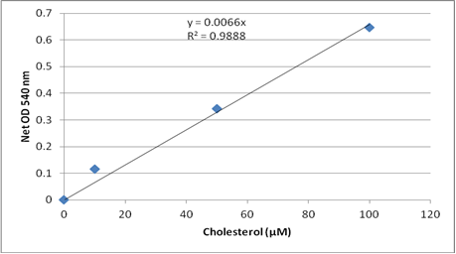
(利用公式算出未知樣品濃度)
10.For the conversion of results from μM to mg/dl, divide the cholesterol concentration(μM) by 25.9.
(將算出的μM的濃度除以25.9換算成mg/dl的濃度,50μM=1.93 mg/dl)
III. References
1. Admundson, D.M., et al. (1999) J. Biochem. Biophys. Meth. 38: 43-52.
2. Cholesterol and Triglyceride concentrations in Serum/Plasma Pairs. (1977) Clin. Chem. 23: 60-63.
3. Fossati, P., et al. (1982) Clin. Chem. 28: 2077-2080.
4. Ledwozyw, A., et al. (1986) Clin. Chim. Acta. 155: 275-284.
5. Lee, S.M. et al. (2008) Lipids 43: 419-429.
6. Cell Biolabs, Inc.-Total Cholesterol Assay Kit (Colorimetric),Catalog Number:STA-384
Video of Experimental Procedures:https://youtu.be/oR4w8wLS_VE
Thumbnail Boris TM Wikipedia C0

