6.2: The Viral Life Cycle
- Page ID
- 23630
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Skills to Develop
- Describe the lytic and lysogenic life cycles
- Describe the replication process of animal viruses
- Describe unique characteristics of retroviruses and latent viruses
- Discuss human viruses and their virus-host cell interactions
- Explain the process of transduction
- Describe the replication process of plant viruses
All viruses depend on cells for reproduction and metabolic processes. By themselves, viruses do not encode for all of the enzymes necessary for viral replication. But within a host cell, a virus can commandeer cellular machinery to produce more viral particles. Bacteriophages replicate only in the cytoplasm, since prokaryotic cells do not have a nucleus or organelles. In eukaryotic cells, most DNA viruses can replicate inside the nucleus, with an exception observed in the large DNA viruses, such as the poxviruses, that can replicate in the cytoplasm. RNA viruses that infect animal cells often replicate in the cytoplasm.
The Life Cycle of Viruses with Prokaryote Hosts
The life cycle of bacteriophages has been a good model for understanding how viruses affect the cells they infect, since similar processes have been observed for eukaryotic viruses, which can cause immediate death of the cell or establish a latent or chronic infection. Virulent phages typically lead to the death of the cell through cell lysis. Temperate phages, on the other hand, can become part of a host chromosome and are replicated with the cell genome until such time as they are induced to make newly assembled viruses, or progeny viruses.
The Lytic Cycle
During the lytic cycle of virulent phage, the bacteriophage takes over the cell, reproduces new phages, and destroys the cell. T-even phage is a good example of a well-characterized class of virulent phages. There are five stages in the bacteriophage lytic cycle (see Figure \(\PageIndex{1}\)). Attachment is the first stage in the infection process in which the phage interacts with specific bacterial surface receptors (e.g., lipopolysaccharides and OmpC protein on host surfaces). Most phages have a narrow host range and may infect one species of bacteria or one strain within a species. This unique recognition can be exploited for targeted treatment of bacterial infection by phage therapy or for phage typing to identify unique bacterial subspecies or strains. The second stage of infection is entry or penetration. This occurs through contraction of the tail sheath, which acts like a hypodermic needle to inject the viral genome through the cell wall and membrane. The phage head and remaining components remain outside the bacteria.
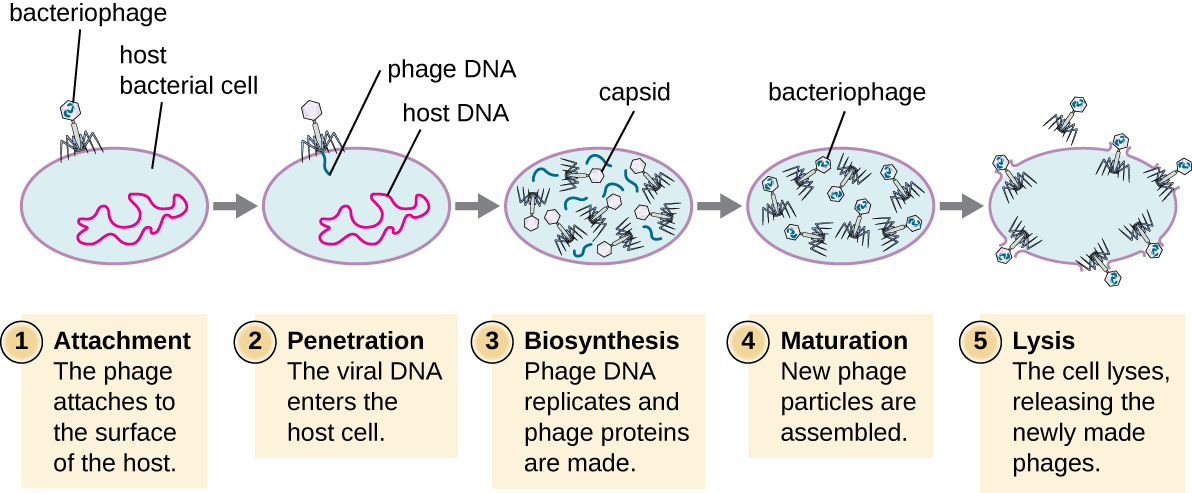
Figure \(\PageIndex{1}\): A virulent phage shows only the lytic cycle pictured here. In the lytic cycle, the phage replicates and lyses the host cell.
The third stage of infection is biosynthesis of new viral components. After entering the host cell, the virus synthesizes virus-encoded endonucleases to degrade the bacterial chromosome. It then hijacks the host cell to replicate, transcribe, and translate the necessary viral components (capsomeres, sheath, base plates, tail fibers, and viral enzymes) for the assembly of new viruses. Polymerase genes are usually expressed early in the cycle, while capsid and tail proteins are expressed later. During the maturation phase, new virions are created. To liberate free phages, the bacterial cell wall is disrupted by phage proteins such as holin or lysozyme. The final stage is release. Mature viruses burst out of the host cell in a process called lysis and the progeny viruses are liberated into the environment to infect new cells.
The Lysogenic Cycle
In a lysogenic cycle, the phage genome also enters the cell through attachment and penetration. A prime example of a phage with this type of life cycle is the lambda phage. During the lysogenic cycle, instead of killing the host, the phage genome integrates into the bacterial chromosome and becomes part of the host. The integrated phage genome is called a prophage. A bacterial host with a prophage is called a lysogen. The process in which a bacterium is infected by a temperate phage is called lysogeny. It is typical of temperate phages to be latent or inactive within the cell. As the bacterium replicates its chromosome, it also replicates the phage’s DNA and passes it on to new daughter cells during reproduction. The presence of the phage may alter the phenotype of the bacterium, since it can bring in extra genes (e.g., toxin genes that can increase bacterial virulence). This change in the host phenotype is called lysogenic conversion or phage conversion. Some bacteria, such as Vibrio cholerae and Clostridium botulinum, are less virulent in the absence of the prophage. The phages infecting these bacteria carry the toxin genes in their genome and enhance the virulence of the host when the toxin genes are expressed. In the case of V. cholera, phage encoded toxin can cause severe diarrhea; in C. botulinum, the toxin can cause paralysis. During lysogeny, the prophage will persist in the host chromosome until induction, which results in the excision of the viral genome from the host chromosome. After induction has occurred the temperate phage can proceed through a lytic cycle and then undergo lysogeny in a newly infected cell (see Figure \(\PageIndex{2}\)).

Figure \(\PageIndex{2}\): A temperate bacteriophage has both lytic and lysogenic cycles. In the lysogenic cycle, phage DNA is incorporated into the host genome, forming a prophage, which is passed on to subsequent generations of cells. Environmental stressors such as starvation or exposure to toxic chemicals may cause the prophage to be excised and enter the lytic cycle.
This video illustrates the stages of the lysogenic life cycle of a bacteriophage and the transition to a lytic phase.
Exercise \(\PageIndex{1}\)
Is a latent phage undetectable in a bacterium?
Transduction
Transduction occurs when a bacteriophage transfers bacterial DNA from one bacterium to another during sequential infections. There are two types of transduction: generalized and specialized transduction. During the lytic cycle of viral replication, the virus hijacks the host cell, degrades the host chromosome, and makes more viral genomes. As it assembles and packages DNA into the phage head, packaging occasionally makes a mistake. Instead of packaging viral DNA, it takes a random piece of host DNA and inserts it into the capsid. Once released, this virion will then inject the former host’s DNA into a newly infected host. The asexual transfer of genetic information can allow for DNA recombination to occur, thus providing the new host with new genes (e.g., an antibiotic-resistance gene, or a sugar-metabolizing gene). Generalized transduction occurs when a random piece of bacterial chromosomal DNA is transferred by the phage during the lytic cycle. Specialized transduction occurs at the end of the lysogenic cycle, when the prophage is excised and the bacteriophage enters the lytic cycle. Since the phage is integrated into the host genome, the prophage can replicate as part of the host. However, some conditions (e.g., ultraviolet light exposure or chemical exposure) stimulate the prophage to undergo induction, causing the phage to excise from the genome, enter the lytic cycle, and produce new phages to leave host cells. During the process of excision from the host chromosome, a phage may occasionally remove some bacterial DNA near the site of viral integration. The phage and host DNA from one end or both ends of the integration site are packaged within the capsid and are transferred to the new, infected host. Since the DNA transferred by the phage is not randomly packaged but is instead a specific piece of DNA near the site of integration, this mechanism of gene transfer is referred to as specialized transduction (see Figure \(\PageIndex{3}\)). The DNA can then recombine with host chromosome, giving the latter new characteristics. Transduction seems to play an important role in the evolutionary process of bacteria, giving them a mechanism for asexual exchange of genetic information.
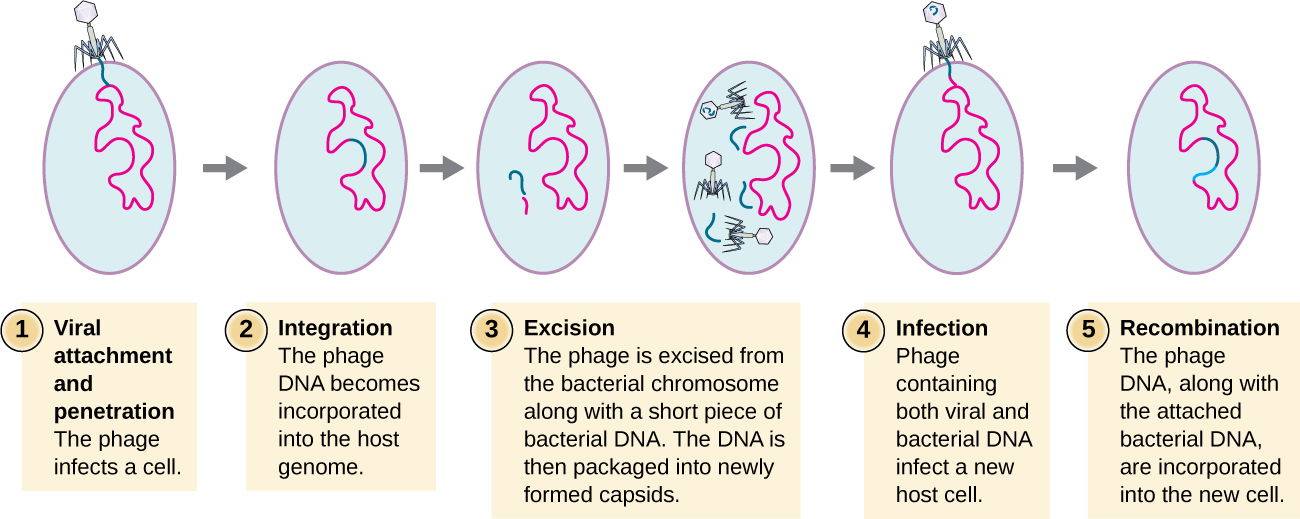
Figure \(\PageIndex{3}\): This flowchart illustrates the mechanism of specialized transduction. An integrated phage excises, bringing with it a piece of the DNA adjacent to its insertion point. On reinfection of a new bacterium, the phage DNA integrates along with the genetic material acquired from the previous host.
Exercise \(\PageIndex{2}\)
Which phage life cycle is associated with which forms of transduction?
Life Cycle of Viruses with Animal Hosts
Lytic animal viruses follow similar infection stages to bacteriophages: attachment, penetration, biosynthesis, maturation, and release (see Figure \(\PageIndex{4}\)). However, the mechanisms of penetration, nucleic-acid biosynthesis, and release differ between bacterial and animal viruses. After binding to host receptors, animal viruses enter through endocytosis(engulfment by the host cell) or through membrane fusion (viral envelope with the host cell membrane). Many viruses are host specific, meaning they only infect a certain type of host; and most viruses only infect certain types of cells within tissues. This specificity is called a tissue tropism. Examples of this are demonstrated by the poliovirus, which exhibits tropism for the tissues of the brain and spinal cord, or the influenza virus, which has a primary tropism for the respiratory tract.
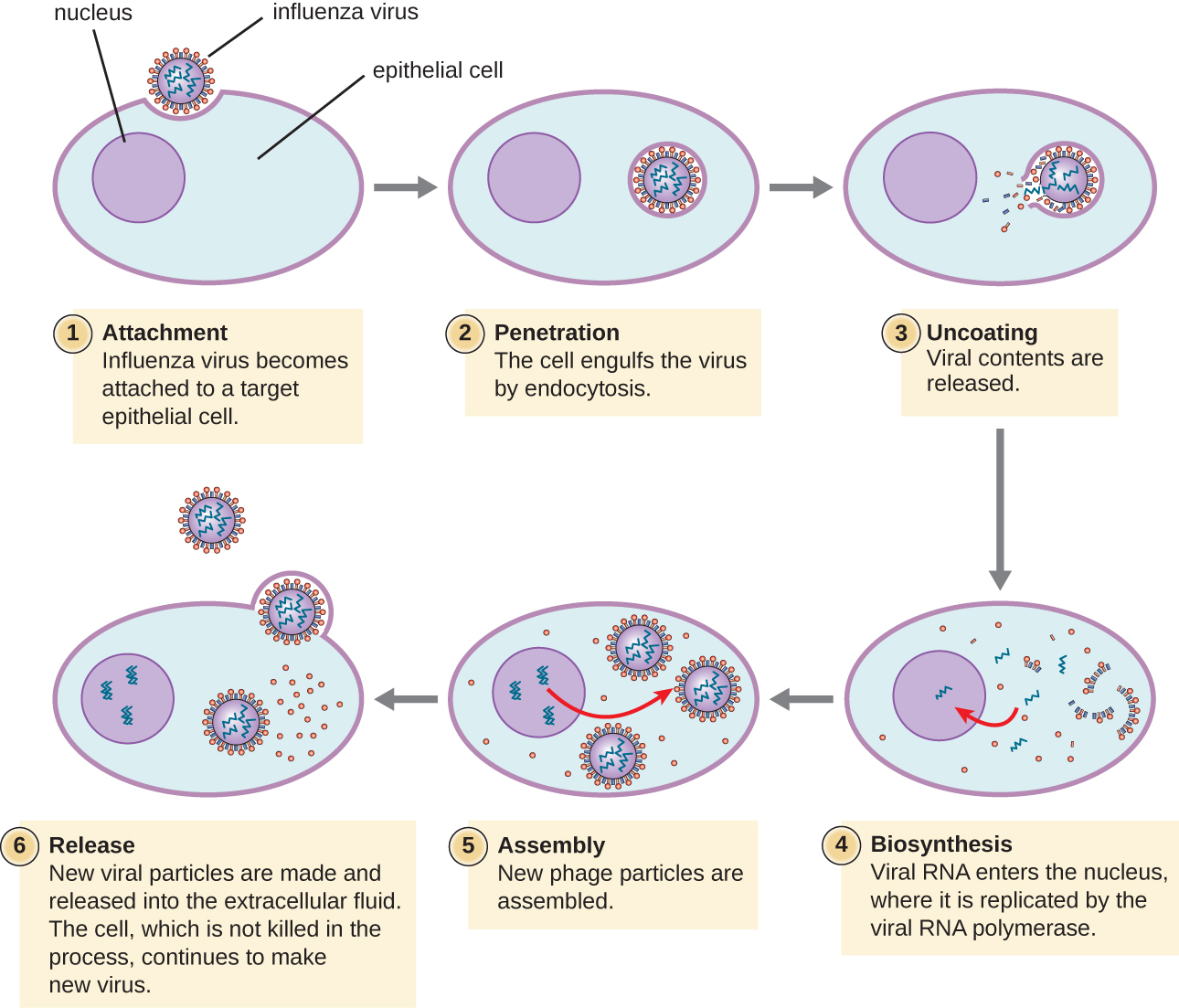
Figure \(\PageIndex{4}\): In influenza virus infection, viral glycoproteins attach the virus to a host epithelial cell. As a result, the virus is engulfed. Viral RNA and viral proteins are made and assembled into new virions that are released by budding.
Animal viruses do not always express their genes using the normal flow of genetic information—from DNA to RNA to protein. Some viruses have a dsDNA genome like cellular organisms and can follow the normal flow. However, others may have ssDNA, dsRNA, or ssRNA genomes. The nature of the genome determines how the genome is replicated and expressed as viral proteins. If a genome is ssDNA, host enzymes will be used to synthesize a second strand that is complementary to the genome strand, thus producing dsDNA. The dsDNA can now be replicated, transcribed, and translated similar to host DNA.
If the viral genome is RNA, a different mechanism must be used. There are three types of RNA genome: dsRNA, positive (+) single-strand (+ssRNA) or negative (−) single-strand RNA (−ssRNA). If a virus has a +ssRNA genome, it can be translated directly to make viral proteins. Viral genomic +ssRNA acts like cellular mRNA. However, if a virus contains a −ssRNA genome, the host ribosomes cannot translate it until the −ssRNA is replicated into +ssRNA by viral RNA-dependent RNA polymerase (RdRP) (see Figure \(\PageIndex{5}\)). The RdRP is brought in by the virus and can be used to make +ssRNA from the original −ssRNA genome. The RdRP is also an important enzyme for the replication of dsRNA viruses, because it uses the negative strand of the double-stranded genome as a template to create +ssRNA. The newly synthesized +ssRNA copies can then be translated by cellular ribosomes.
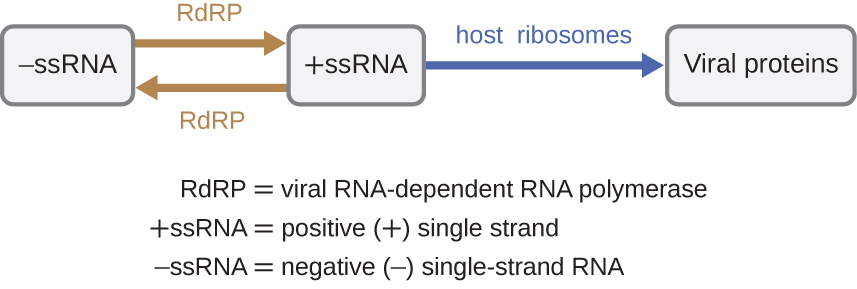
Figure \(\PageIndex{5}\): RNA viruses can contain +ssRNA that can be directly read by the ribosomes to synthesize viral proteins. Viruses containing −ssRNA must first use the −ssRNA as a template for the synthesis of +ssRNA before viral proteins can be synthesized.
An alternative mechanism for viral nucleic acid synthesis is observed in the retroviruses, which are +ssRNA viruses (see Figure \(\PageIndex{6}\)). Single-stranded RNA viruses such as HIV carry a special enzyme called reverse transcriptase within the capsid that synthesizes a complementary ssDNA (cDNA) copy using the +ssRNA genome as a template. The ssDNA is then made into dsDNA, which can integrate into the host chromosome and become a permanent part of the host. The integrated viral genome is called a provirus. The virus now can remain in the host for a long time to establish a chronic infection. The provirus stage is similar to the prophage stage in a bacterial infection during the lysogenic cycle. However, unlike prophage, the provirus does not undergo excision after splicing into the genome.

Figure \(\PageIndex{6}\): HIV, an enveloped, icosahedral retrovirus, attaches to a cell surface receptor of an immune cell and fuses with the cell membrane. Viral contents are released into the cell, where viral enzymes convert the single-stranded RNA genome into DNA and incorporate it into the host genome. (credit: modification of work by NIAID, NIH)
Exercise \(\PageIndex{3}\)
Is RNA-dependent RNA polymerase made from a viral gene or a host gene?
Persistent Infections
Persistent infection occurs when a virus is not completely cleared from the system of the host but stays in certain tissues or organs of the infected person. The virus may remain silent or undergo productive infection without seriously harming or killing the host. Mechanisms of persistent infection may involve the regulation of the viral or host gene expressions or the alteration of the host immune response. The two primary categories of persistent infections are latent infection and chronic infection. Examples of viruses that cause latent infections include herpes simplex virus (oral and genital herpes), varicella-zoster virus (chickenpox and shingles), and Epstein-Barr virus (mononucleosis). Hepatitis C virus and HIV are two examples of viruses that cause long-term chronic infections.
Latent Infection
Not all animal viruses undergo replication by the lytic cycle. There are viruses that are capable of remaining hidden or dormant inside the cell in a process called latency. These types of viruses are known as latent viruses and may cause latent infections. Viruses capable of latency may initially cause an acute infection before becoming dormant.
For example, the varicella-zoster virus infects many cells throughout the body and causes chickenpox, characterized by a rash of blisters covering the skin. About 10 to 12 days postinfection, the disease resolves and the virus goes dormant, living within nerve-cell ganglia for years. During this time, the virus does not kill the nerve cells or continue replicating. It is not clear why the virus stops replicating within the nerve cells and expresses few viral proteins but, in some cases, typically after many years of dormancy, the virus is reactivated and causes a new disease called shingles (Figure \(\PageIndex{7}\)). Whereas chickenpox affects many areas throughout the body, shingles is a nerve cell-specific disease emerging from the ganglia in which the virus was dormant.
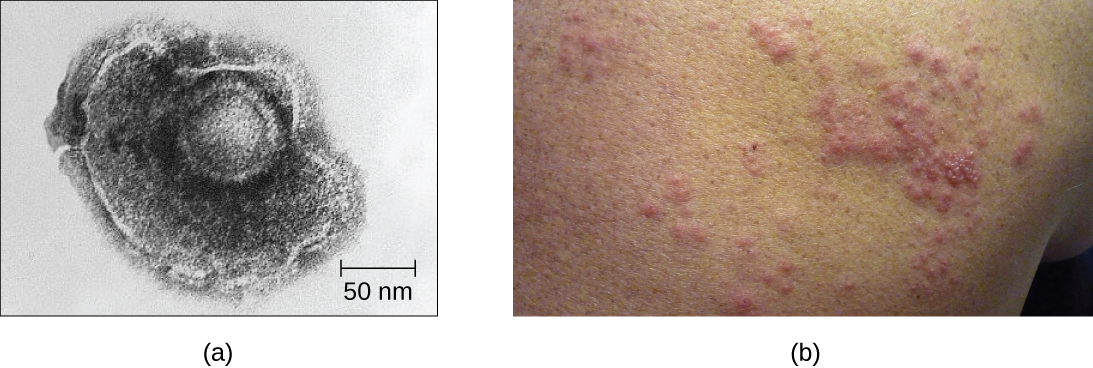
Figure \(\PageIndex{7}\): (a) Varicella-zoster, the virus that causes chickenpox, has an enveloped icosahedral capsid visible in this transmission electron micrograph. Its double-stranded DNA genome becomes incorporated in the host DNA. (b) After a period of latency, the virus can reactivate in the form of shingles, usually manifesting as a painful, localized rash on one side of the body. (credit a: modification of work by Erskine Palmer and B.G. Partin—scale-bar data from Matt Russell; credit b: modification of work by Rosmarie Voegtli)
Latent viruses may remain dormant by existing as circular viral genome molecules outside of the host chromosome. Others become proviruses by integrating into the host genome. During dormancy, viruses do not cause any symptoms of disease and may be difficult to detect. A patient may be unaware that he or she is carrying the virus unless a viral diagnostic test has been performed.
Chronic Infection
A chronic infection is a disease with symptoms that are recurrent or persistent over a long time. Some viral infections can be chronic if the body is unable to eliminate the virus. HIV is an example of a virus that produces a chronic infection, often after a long period of latency. Once a person becomes infected with HIV, the virus can be detected in tissues continuously thereafter, but untreated patients often experience no symptoms for years. However, the virus maintains chronic persistence through several mechanisms that interfere with immune function, including preventing expression of viral antigens on the surface of infected cells, altering immune cells themselves, restricting expression of viral genes, and rapidly changing viral antigens through mutation. Eventually, the damage to the immune system results in progression of the disease leading to acquired immunodeficiency syndrome (AIDS). The various mechanisms that HIV uses to avoid being cleared by the immune system are also used by other chronically infecting viruses, including the hepatitis C virus.
Exercise \(\PageIndex{4}\)
In what two ways can a virus manage to maintain a persistent infection?
Life Cycle of Viruses with Plant Hosts
Plant viruses are more similar to animal viruses than they are to bacteriophages. Plant viruses may be enveloped or non-enveloped. Like many animal viruses, plant viruses can have either a DNA or RNA genome and be single stranded or double stranded. However, most plant viruses do not have a DNA genome; the majority have a +ssRNA genome, which acts like messenger RNA (mRNA). Only a minority of plant viruses have other types of genomes.
Plant viruses may have a narrow or broad host range. For example, the citrus tristeza virus infects only a few plants of the Citrus genus, whereas the cucumber mosaic virus infects thousands of plants of various plant families. Most plant viruses are transmitted by contact between plants, or by fungi, nematodes, insects, or other arthropods that act as mechanical vectors. However, some viruses can only be transferred by a specific type of insect vector; for example, a particular virus might be transmitted by aphids but not whiteflies. In some cases, viruses may also enter healthy plants through wounds, as might occur due to pruning or weather damage.
Viruses that infect plants are considered biotrophic parasites, which means that they can establish an infection without killing the host, similar to what is observed in the lysogenic life cycles of bacteriophages. Viral infection can be asymptomatic (latent) or can lead to cell death (lytic infection). The life cycle begins with the penetration of the virus into the host cell. Next, the virus is uncoated within the cytoplasm of the cell when the capsid is removed. Depending on the type of nucleic acid, cellular components are used to replicate the viral genome and synthesize viral proteins for assembly of new virions. To establish a systemic infection, the virus must enter a part of the vascular system of the plant, such as the phloem. The time required for systemic infection may vary from a few days to a few weeks depending on the virus, the plant species, and the environmental conditions. The virus life cycle is complete when it is transmitted from an infected plant to a healthy plant.
Exercise \(\PageIndex{5}\)
What is the structure and genome of a typical plant virus?
Viral Growth Curve
Unlike the growth curve for a bacterial population, the growth curve for a virus population over its life cycle does not follow a sigmoidal curve. During the initial stage, an inoculum of virus causes infection. In the eclipse phase, viruses bind and penetrate the cells with no virions detected in the medium. The chief difference that next appears in the viral growth curve compared to a bacterial growth curve occurs when virions are released from the lysed host cell at the same time. Such an occurrence is called a burst, and the number of virions per bacterium released is described as the burst size. In a one-step multiplication curve for bacteriophage, the host cells lyse, releasing many viral particles to the medium, which leads to a very steep rise in viral titer (the number of virions per unit volume). If no viable host cells remain, the viral particles begin to degrade during the decline of the culture (see Figure \(\PageIndex{8}\)).
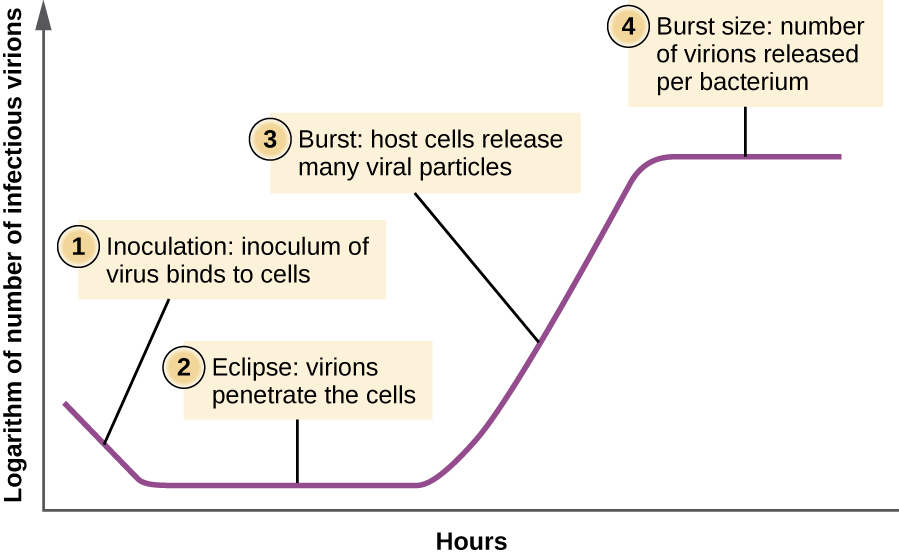
Figure \(\PageIndex{8}\): The one-step multiplication curve for a bacteriophage population follows three steps: 1) inoculation, during which the virions attach to host cells; 2) eclipse, during which entry of the viral genome occurs; and 3) burst, when sufficient numbers of new virions are produced and emerge from the host cell. The burst size is the maximum number of virions produced per bacterium.
Exercise \(\PageIndex{6}\)
What aspect of the life cycle of a virus leads to the sudden increase in the growth curve?
unregistered treatments
Ebola is incurable and deadly. The outbreak in West Africa in 2014 was unprecedented, dwarfing other human Ebola epidemics in the level of mortality. Of 24,666 suspected or confirmed cases reported, 10,179 people died.1
No approved treatments or vaccines for Ebola are available. While some drugs have shown potential in laboratory studies and animal models, they have not been tested in humans for safety and effectiveness. Not only are these drugs untested or unregistered but they are also in short supply.
Given the great suffering and high mortality rates, it is fair to ask whether unregistered and untested medications are better than none at all. Should such drugs be dispensed and, if so, who should receive them, in light of their extremely limited supplies? Is it ethical to treat untested drugs on patients with Ebola? On the other hand, is it ethical to withhold potentially life-saving drugs from dying patients? Or should the drugs perhaps be reserved for health-care providers working to contain the disease?
In August 2014, two infected US aid workers and a Spanish priest were treated with ZMapp, an unregistered drug that had been tested in monkeys but not in humans. The two American aid workers recovered, but the priest died. Later that month, the WHO released a report on the ethics of treating patients with the drug. Since Ebola is often fatal, the panel reasoned that it is ethical to give the unregistered drugs and unethical to withhold them for safety concerns. This situation is an example of “compassionate use” outside the well-established system of regulation and governance of therapies.
ebola in the us
On September 24, 2014, Thomas Eric Duncan arrived at the Texas Health Presbyterian Hospital in Dallas complaining of a fever, headache, vomiting, and diarrhea—symptoms commonly observed in patients with the cold or the flu. After examination, an emergency department doctor diagnosed him with sinusitis, prescribed some antibiotics, and sent him home. Two days later, Duncan returned to the hospital by ambulance. His condition had deteriorated and additional blood tests confirmed that he has been infected with the Ebola virus.
Further investigations revealed that Duncan had just returned from Liberia, one of the countries in the midst of a severe Ebola epidemic. On September 15, nine days before he showed up at the hospital in Dallas, Duncan had helped transport an Ebola-stricken neighbor to a hospital in Liberia. The hospital continued to treat Duncan, but he died several days after being admitted.
The timeline of the Duncan case is indicative of the life cycle of the Ebola virus. The incubation time for Ebola ranges from 2 days to 21 days. Nine days passed between Duncan’s exposure to the virus infection and the appearance of his symptoms. This corresponds, in part, to the eclipse period in the growth of the virus population. During the eclipse phase, Duncan would have been unable to transmit the disease to others. However, once an infected individual begins exhibiting symptoms, the disease becomes very contagious. Ebola virus is transmitted through direct contact with droplets of bodily fluids such as saliva, blood, and vomit. Duncan could conceivably have transmitted the disease to others at any time after he began having symptoms, presumably some time before his arrival at the hospital in Dallas. Once a hospital realizes a patient like Duncan is infected with Ebola virus, the patient is immediately quarantined, and public health officials initiate a back trace to identify everyone with whom a patient like Duncan might have interacted during the period in which he was showing symptoms.
Public health officials were able to track down 10 high-risk individuals (family members of Duncan) and 50 low-risk individuals to monitor them for signs of infection. None contracted the disease. However, one of the nurses charged with Duncan’s care did become infected. This, along with Duncan’s initial misdiagnosis, made it clear that US hospitals needed to provide additional training to medical personnel to prevent a possible Ebola outbreak in the US.
Exercise \(\PageIndex{7}\)
- What types of training can prepare health professionals to contain emerging epidemics like the Ebola outbreak of 2014?
- What is the difference between a contagious pathogen and an infectious pathogen?

Figure \(\PageIndex{9}\): Researchers working with Ebola virus use layers of defenses against accidental infection, including protective clothing, breathing systems, and negative air-pressure cabinets for bench work. (credit: modification of work by Randal J. Schoepp)
For additional information about Ebola, please visit the CDC website.
Summary
- Many viruses target specific hosts or tissues. Some may have more than one host.
- Many viruses follow several stages to infect host cells. These stages include attachment, penetration, uncoating, biosynthesis, maturation, and release.
- Bacteriophages have a lytic or lysogenic cycle. The lytic cycle leads to the death of the host, whereas the lysogenic cycle leads to integration of phage into the host genome.
- Bacteriophages inject DNA into the host cell, whereas animal viruses enter by endocytosis or membrane fusion.
- Animal viruses can undergo latency, similar to lysogeny for a bacteriophage.
- The majority of plant viruses are positive-strand ssRNA and can undergo latency, chronic, or lytic infection, as observed for animal viruses.
- The growth curve of bacteriophage populations is a one-step multiplication curve and not a sigmoidal curve, as compared to the bacterial growth curve.
- Bacteriophages transfer genetic information between hosts using either generalized or specialized transduction.
Footnotes
- 1 World Health Organization. “WHO Ebola Data and Statistics.” March 18, 2005. http://apps.who.int/gho/data/view.eb...150318?lang=en
Contributor
Nina Parker, (Shenandoah University), Mark Schneegurt (Wichita State University), Anh-Hue Thi Tu (Georgia Southwestern State University), Philip Lister (Central New Mexico Community College), and Brian M. Forster (Saint Joseph’s University) with many contributing authors. Original content via Openstax (CC BY 4.0; Access for free at https://openstax.org/books/microbiology/pages/1-introduction)


