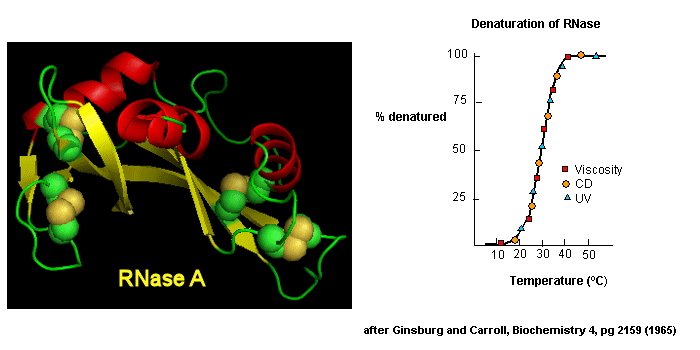
Early studies of protein folding involved small proteins which could be denatured and refolded in a reversible fashion. A two state model, D <===> N, was assumed. The denaturants were heat, urea, or guanidine HCl. Since the denatured states are less compact than the native state, the viscosity of the solution can be used as a measure of denaturation/renaturation. Likewise, the amino acid side chains in the differing states would be in different environments. The aromatic amino acid Trp, Phe, and Tyr absorb UV light. After excitation, the electrons decay to the ground state through several processes. Some vibrational relaxation occurs, bringing the electrons to lower vibrational energy levels. Some of the electrons can then fall to various vibrational levels at lower principle energy states through a radiative process. The photons emitted are lower in energy and hence longer in wavelength. The emitted light is termed fluorescence. The wavelength of maximum fluorescent intensity and the lifetime of the fluorescence decay is very sensitive to the environment of the amino acids. Hence fluorescence can also be used to measure changes in protein conformation. Other spectral techniques like CD spectroscopy as well as simple absorbance measurements, are used. For small, single domain proteins (such as RNase) undergoing reversible denatuation, graphs showing the extent of denaturation using each technique above, are superimposable, giving strong validity to the two state model.
Figure: Reversible denaturation
Proteins that fold without easily discernable, long lived intermediates and following a simple two state model, D <=> N are said to undergo cooperative folding. This simple model needed to be expanded as more proteins were studied. Some intermediates in the process were detected.
- Some proteins show two steps, one slow, one quick, in refolding studies, suggesting an intermediate. The longer a protein is kept in the denatured state, the more likely it is to display an intermediate. One accepted explanation for this phenomena is that during an extended time in the D state, some X-Pro bonds might isomerize from trans to the cis state, to form an intermediate. Alternatively, as in the case of RNase, which has a cis X-Pro bond in the native state, denaturation causes an isomerization to the trans state. In the case of RNase, to refold, the accumulating intermediate I must reisomerize in a slow step to the cis state, followed by a quick return to the N state.
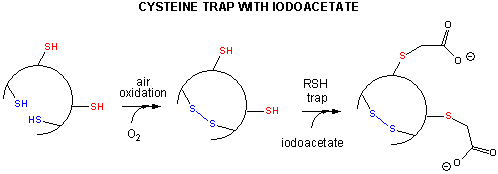
- Some proteins which contain multiple disulfide bonds that must reform correctly after reductive denaturation can refold into intermediates with the wrong S-S partner. Such intermediates can be trapped by stopping further S-S formation during refolding with the addition of iodoacetamide.
Figure: addition of iodoacetamide
As an example consider the following data on bovine pancreatic trypsin inhibitor.
Figure: Bovine pancreatic trypsin inhbitor (BPTI): Folding Kinetics - only native disulfide structures seem to form.
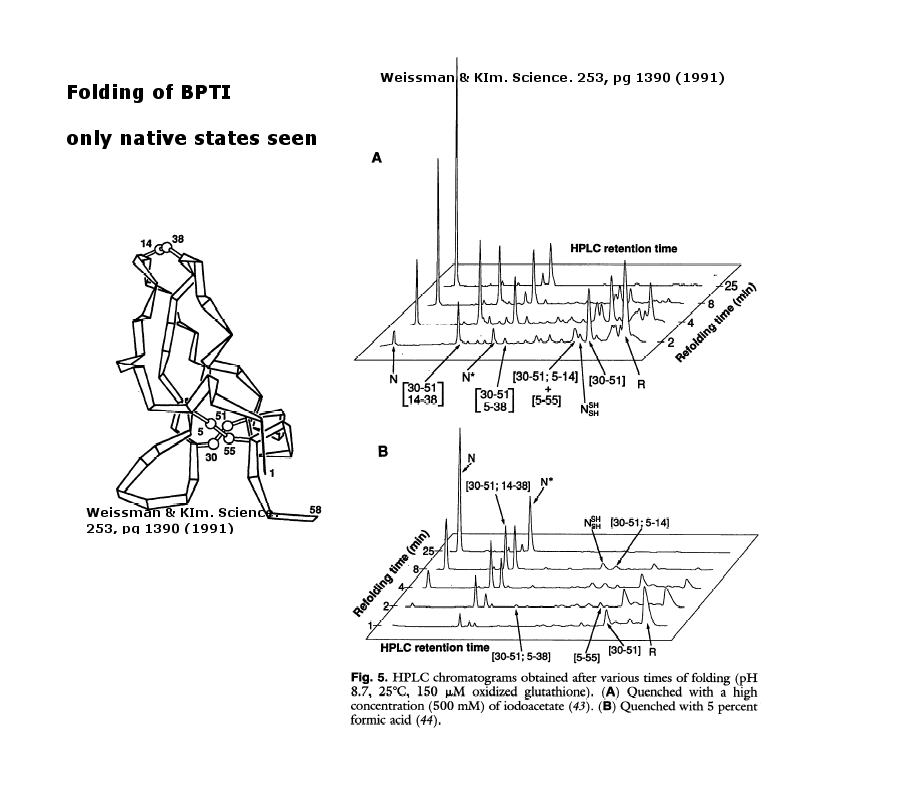
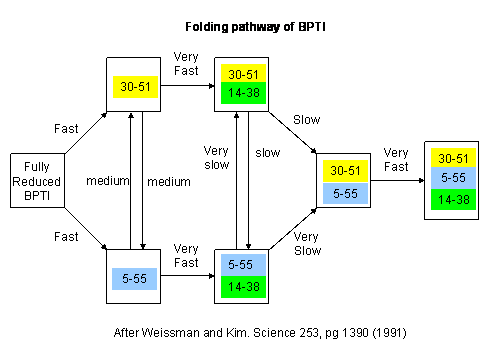
Figure: BPTI Folding Pathway In Vitro - gives possible scheme of folding intermediates
- Some proteins form partially folded but stable intermediates when folded under partially denaturing conditions. A good example is lactalbumin, which under mildly acidic conditions (pH 4), low levels of guanidine HCl, or neutral pH and low ionic strength in the absence of calcium (which normally binds to the protein), forms a stable, isolatable intermediate (I) called the molten globule (MG). The image below shows the folded state with two calcium ions bound.
Figure: lactalbumin (image made with Pymol)
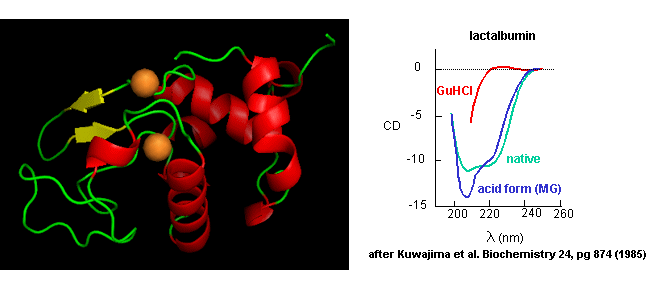
Data show that the MG is about 50% larger in volume than the N state. This compares to the denatured state, which can be 300% larger than the native state. Hence, it is more like the native state as studied by hydrodynamic techniques, but with more solvent accessibility of hydrophobic side chains. The MG has a similar CD spectra as the native state, but the aromatic side chains display the same UV absorption and fluorescent characteristics as the protein in 6 M guanidine HCl, suggesting that the final tertiary state has not yet completely formed. The secondary structure in the MG may not be the same as in the native state
NMR can also be used to detect folding intermediates. Using this technique, proteins are unfolded in D2O, which will cause the exchange of all Cs with ionizable protons, including, the amide Hs. An amine is a weak base (pKb around 3.5) so its conjugate acid, the protonated amine, has a pKa of around 9.5. An amide or peptide bond would be a weaker base than an amine since it's lone pair is less available (due to delocalization through resonance) for sharing with a proton. The pKa for the conjugate acid of the amide (in which the amide N is protonated and has a plus charge) is much lower, around -0.5, than the pKa for the conjugate acid of an amine. At 2 pH units greater than its pKa, the charged amide N is close to 100% deprotonated The pka of the protonated group is important since the rate of H exchange is related to the pKa, holding other variables constant. The pka of an unprotonated amine (RNH2 -> RNH- is very high (30s) and hence deprotonation of the RNH2 amine to form RNH- is not likely under normal conditions.
Figure: Exchange of all Cs with ionizable protons, including the amide Hs
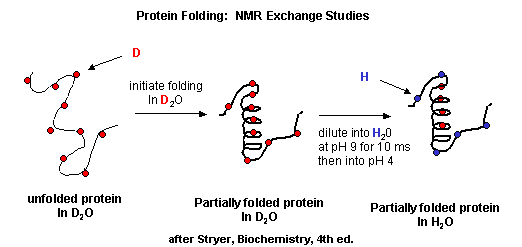
Refolding is initiated by diluting the protein into a solution without the denaturatant, but still in D2O. As the protein folds and becomes more compact, the buried atoms are now sequestered from the solvent, and no longer readily exchange Ds. Then the protein is placed in H2O at pH 9.0 for 10 ms, after which the pH is changed to pH 4.0. D --> H exchange is promoted at high pH, and quenched for the amide Ds and Hs at low pH. Amide H's that continue to exchange must be accessible to water. Those that aren't are usually buried in secondary structure.
- Hydrogen Exchange and Mass Spectrometry in the Study of Proteins
Figure: Experimental data on model proteins. How would you interpret these graphs.
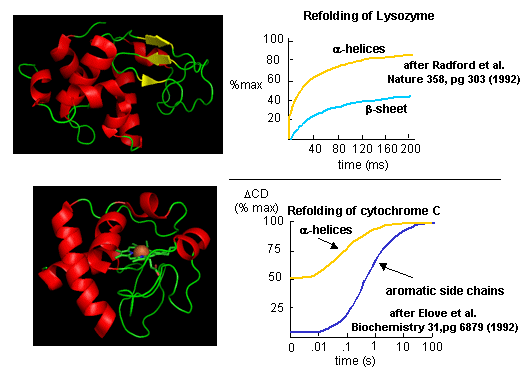
When the same techniques are applied to large, multidomain or oligomeric proteins, only a few percent refold in vitro. Incorrect intermolecular interactions and heterogeneous aggregation seems to be the main problems which prevent correct protein folding in vitro.
 Jmol: Updated Apolactalbumin (w/o Ca2+)/Hololactalbumin Jmol14 (Java) | JSMol (HTML5)
Jmol: Updated Apolactalbumin (w/o Ca2+)/Hololactalbumin Jmol14 (Java) | JSMol (HTML5)
Contributors and Attributions









