8.3: DNA Replication
- Page ID
- 35709
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)What you’ll learn to do: Explain the role of complementary base pairing in the precise replication process of DNA
In this outcome, we’ll learn more about the precise structure of DNA and how it replicates. Watch this video for a quick introduction to this topic:
- Outline the basic steps in DNA replication
- Identify the major enzymes that play a role in DNA replication
- Identify the key proofreading processes in DNA replication
Basics of DNA Replication
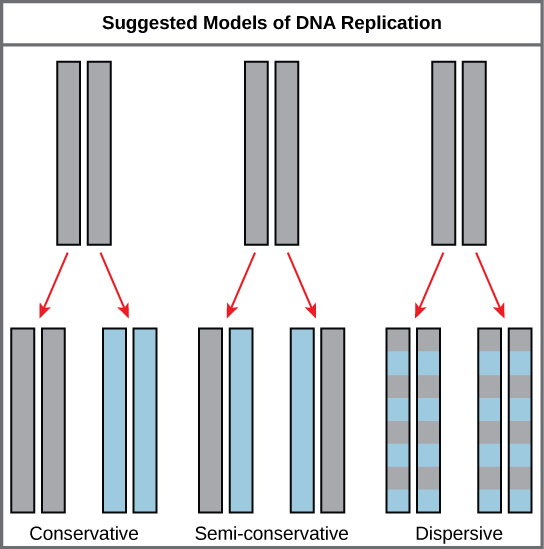
The elucidation of the structure of the double helix provided a hint as to how DNA divides and makes copies of itself. This model suggests that the two strands of the double helix separate during replication, and each strand serves as a template from which the new complementary strand is copied. What was not clear was how the replication took place. There were three models suggested: conservative, semi-conservative, and dispersive (see Figure 1).
In conservative replication, the parental DNA remains together, and the newly formed daughter strands are together. The semi-conservative method suggests that each of the two parental DNA strands act as a template for new DNA to be synthesized; after replication, each double-stranded DNA includes one parental or “old” strand and one “new” strand. In the dispersive model, both copies of DNA have double-stranded segments of parental DNA and newly synthesized DNA interspersed.
Meselson and Stahl were interested in understanding how DNA replicates. They grew E. coli for several generations in a medium containing a “heavy” isotope of nitrogen (15N) that gets incorporated into nitrogenous bases, and eventually into the DNA (Figure 2).
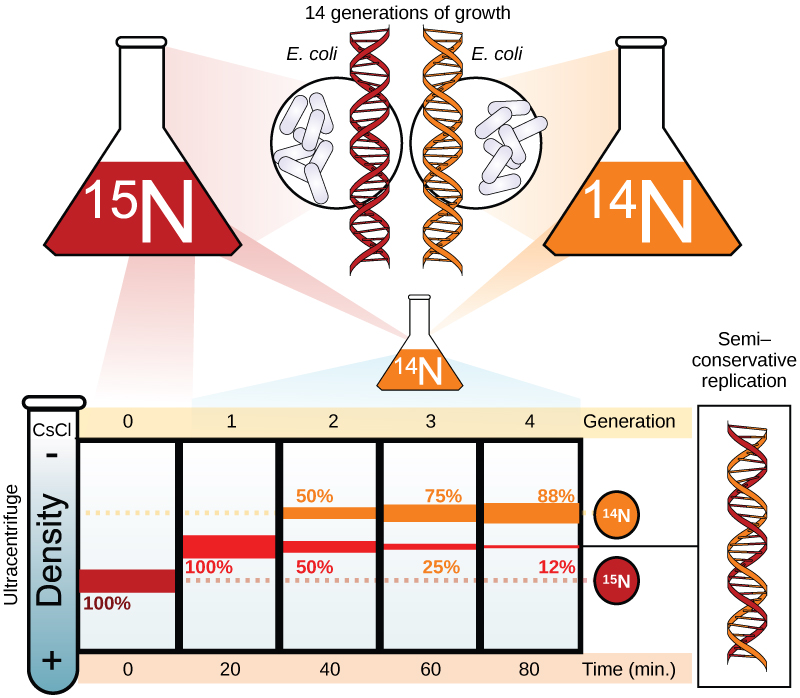
The E. coli culture was then shifted into medium containing 14N and allowed to grow for one generation. The cells were harvested and the DNA was isolated. The DNA was centrifuged at high speeds in an ultracentrifuge. Some cells were allowed to grow for one more life cycle in 14N and spun again. During the density gradient centrifugation, the DNA is loaded into a gradient (typically a salt such as cesium chloride or sucrose) and spun at high speeds of 50,000 to 60,000 rpm. Under these circumstances, the DNA will form a band according to its density in the gradient. DNA grown in 15N will band at a higher density position than that grown in 14N. Meselson and Stahl noted that after one generation of growth in 14N after they had been shifted from 15N, the single band observed was intermediate in position in between DNA of cells grown exclusively in 15N and 14N. This suggested either a semi-conservative or dispersive mode of replication. The DNA harvested from cells grown for two generations in 14N formed two bands: one DNA band was at the intermediate position between 15N and 14N, and the other corresponded to the band of 14N DNA. These results could only be explained if DNA replicates in a semi-conservative manner. Therefore, the other two modes were ruled out.
During DNA replication, each of the two strands that make up the double helix serves as a template from which new strands are copied. The new strand will be complementary to the parental or “old” strand. When two daughter DNA copies are formed, they have the same sequence and are divided equally into the two daughter cells.
Click through this tutorial on DNA replication.
The model for DNA replication suggests that the two strands of the double helix separate during replication, and each strand serves as a template from which the new complementary strand is copied. In conservative replication, the parental DNA is conserved, and the daughter DNA is newly synthesized. The semi-conservative method suggests that each of the two parental DNA strands acts as template for new DNA to be synthesized; after replication, each double-stranded DNA includes one parental or “old” strand and one “new” strand. The dispersive mode suggested that the two copies of the DNA would have segments of parental DNA and newly synthesized DNA. Experimental evidence showed DNA replication is semi-conservative.
Major Enzymes
The process of DNA replication is catalyzed by a type of enzyme called DNA polymerase (poly meaning many, mer meaning pieces, and –ase meaning enzyme; so an enzyme that attaches many pieces of DNA). During replication, the two DNA strands separate at multiple points along the length of the chromosome. These locations are called origins of replication because replication begins at these points. Observe Figure 1: the double helix of the original DNA molecule separates (blue) and new strands are made to match the separated strands. The result will be two DNA molecules, each containing an old and a new strand. Therefore, DNA replication is called semiconservative. The term semiconservative refers to the fact that half of the original molecule (one of the two strands in the double helix) is “conserved” in the new molecule. The original strand is referred to as the template strand because it provides the information, or template, for the newly synthesized strand.

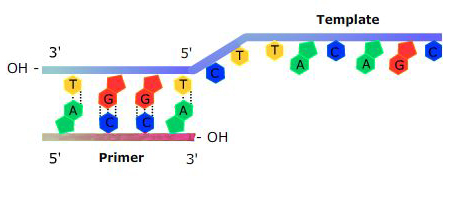
DNA polymerase needs an “anchor” to start adding nucleotides: a short sequence of DNA or RNA that is complementary to the template strand will work to provide a free 3′ end. This sequence is called a primer (Figure 4).
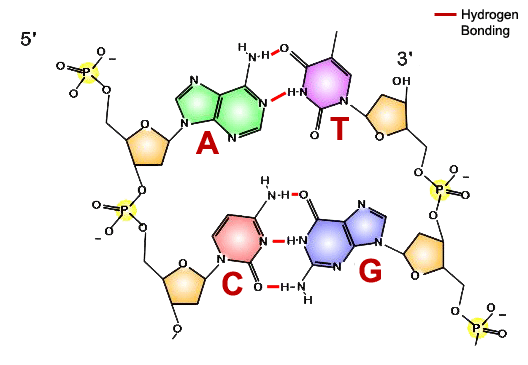
How does DNA polymerase know in what order to add nucleotides? Specific base pairing in DNA is the key to copying the DNA: if you know the sequence of one strand, you can use base pairing rules to build the other strand. Bases form pairs (base pairs) in a very specific way.
True/False: DNA replication requires an enzyme.
- Show Answer
-
True. Most biological reactions rely on the enzyme to speed up the reaction. In the case of DNA replication, this enzyme is DNA polymerase.
What are the building blocks on DNA?
- Deoxyribonucleotides
- Fatty acids
- Ribonucleotides
- Amino acids
- Show Answer
-
Answer a. DNA is a double helix made up of two long chains of deoxyribonucleotides.
True/False: DNA replication requires energy.
- Show Answer
-
True. Making large molecules from small subunits (anabolism) requires energy. What supplies the energy? The building blocks themselves serve as a source of energy. As they get incorporated into the DNA polymer, two phosphate groups are broken off to release energy, some of which is used for making the polymer. Deoxyribonucleotides differ from nucleotides like ATP only by one missing oxygen atom.
We have the building blocks, a source of energy, and a catalyst. What’s missing? We need instruction about the order of nucleotides in the new polymer. Which molecule provides these instructions?
- Protein
- DNA
- Carbohydrate
- Lipid
- Show Answer
-
Answer b. We refer to this DNA as a template. The original information stored in the order of bases will direct the synthesis of the new DNA via base pairing.
There is one more thing required by the DNA polymerase. It cannot just start making a DNA copy of the template strand; it needs a short piece of DNA or RNA with a free hydroxyl group in the right place to attach the nucleotides to. (Remember that synthesis always occurs in one direction—new building blocks are added to the 3′ end.) This component starts the process by giving DNA polymerase something to bind to. What might you call this short piece of nucleic acid?
- A solvent
- A primer
- A converter
- A sealant
- Show Answer
-
Answer b. A primer is used to start this process by giving DNA polymerase something to bind the new nucleotide to.
Now that you understand the basics of DNA replication, we can add a bit of complexity. The two strands of DNA have to be temporarily separated from each other; this job is done by a special enzyme, helicase, that helps unwind and separate the DNA helices (Figure 6). Another issue is that the DNA polymerase only works in one direction along the strand (5′ to 3′), but the double-stranded DNA has two strands oriented in opposite directions. This problem is solved by synthesizing the two strands slightly differently: one new strand grows continuously, the other in bits and pieces. The leading strand grows continuously and the lagging strand is put together with short pieces called Okazaki fragments. These fragments are connected and linked together by an enzyme called DNA ligase. Short fragments of RNA are used as primers for the DNA polymerase.

Which of these separates the two complementary strands of DNA?
- DNA polymerase
- helicase
- RNA primer
- single-strand binding protein
- Show Answer
-
Answer b. Helicase breaks the hydrogen bonds holding together the two strands of DNA.
Which of these attaches complementary bases to the template strand?
- DNA polymerase
- helicase
- RNA primer
- single-strand binding protein
- Show Answer
-
Answer a. DNA polymerase builds the new strand of DNA.
Which of these is later replaced with DNA bases?
- DNA polymerase
- helicase
- RNA primer
- single-strand binding protein
- Show Answers
-
Answer c. the RNA primer is replaced with DNA nucleotides.
Replication in eukaryotes starts at multiple origins of replication. A primer is required to initiate synthesis, which is then extended by DNA polymerase as it adds nucleotides one by one to the growing chain. The leading strand is synthesized continuously, whereas the lagging strand is synthesized in short stretches called Okazaki fragments. The RNA primers are replaced with DNA nucleotides; the DNA remains one continuous strand by linking the DNA fragments with DNA ligase.
Proofreading DNA
DNA replication is a highly accurate process, but mistakes can occasionally occur, such as a DNA polymerase inserting a wrong base. Uncorrected mistakes may sometimes lead to serious consequences, such as cancer. Repair mechanisms correct the mistakes. In rare cases, mistakes are not corrected, leading to mutations; in other cases, repair enzymes are themselves mutated or defective.
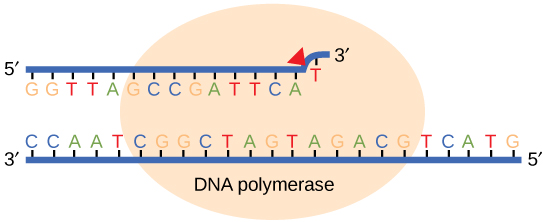
Most of the mistakes during DNA replication are promptly corrected by DNA polymerase by proofreading the base that has been just added (Figure 7). In proofreading, the DNA pol reads the newly added base before adding the next one, so a correction can be made. The polymerase checks whether the newly added base has paired correctly with the base in the template strand. If it is the right base, the next nucleotide is added. If an incorrect base has been added, the enzyme makes a cut at the phosphodiester bond and releases the wrong nucleotide. This is performed by the exonuclease action of DNA pol III. Once the incorrect nucleotide has been removed, a new one will be added again.
Some errors are not corrected during replication, but are instead corrected after replication is completed; this type of repair is known as mismatch repair (Figure 8). The enzymes recognize the incorrectly added nucleotide and excise it; this is then replaced by the correct base. If this remains uncorrected, it may lead to more permanent damage. How do mismatch repair enzymes recognize which of the two bases is the incorrect one? In E. coli, after replication, the nitrogenous base adenine acquires a methyl group; the parental DNA strand will have methyl groups, whereas the newly synthesized strand lacks them. Thus, DNA polymerase is able to remove the wrongly incorporated bases from the newly synthesized, non-methylated strand. In eukaryotes, the mechanism is not very well understood, but it is believed to involve recognition of unsealed nicks in the new strand, as well as a short-term continuing association of some of the replication proteins with the new daughter strand after replication has completed.

In another type of repair mechanism, nucleotide excision repair, enzymes replace incorrect bases by making a cut on both the 3′ and 5′ ends of the incorrect base (Figure 9).
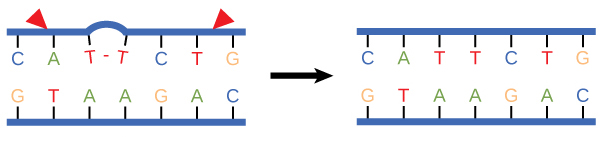
The segment of DNA is removed and replaced with the correctly paired nucleotides by the action of DNA pol. Once the bases are filled in, the remaining gap is sealed with a phosphodiester linkage catalyzed by DNA ligase. This repair mechanism is often employed when UV exposure causes the formation of pyrimidine dimers.
Check Your Understanding
Answer the question(s) below to see how well you understand the topics covered in the previous section. This short quiz does not count toward your grade in the class, and you can retake it an unlimited number of times.
Use this quiz to check your understanding and decide whether to (1) study the previous section further or (2) move on to the next section.
Contributors and Attributions
- Introduction to DNA Replication. Authored by: Shelli Carter and Lumen Learning. Provided by: Lumen Learning. License: CC BY: Attribution
- Biology. Provided by: OpenStax CNX. Located at: http://cnx.org/contents/185cbf87-c72e-48f5-b51e-f14f21b5eabd@10.8. License: CC BY: Attribution. License Terms: Download for free at http://cnx.org/contents/185cbf87-c72...f21b5eabd@10.8
- DNA Replication. Provided by: Open Learning Initiative. Located at: https://oli.cmu.edu/jcourse/workbook/activity/page?context=434a5f7e80020ca6000225da6a4220c9. Project: Introduction to Biology (Open + Free). License: CC BY-NC-SA: Attribution-NonCommercial-ShareAlike
- DNA replication - 3D. Authored by: yourgenome. Located at: https://youtu.be/TNKWgcFPHqw. License: All Rights Reserved. License Terms: Standard YouTube License

