8.4: The Evolution of Aging
- Page ID
- 78369
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)The Evolution of Aging
Senescence or biological aging is the gradual deterioration of functional characteristics in living organisms. The word senescence can refer to either cellular senescence or to senescence of the whole organism. Organismal senescence involves an increase in death rates and/or a decrease in fecundity with increasing age, at least in the latter part of an organism's life cycle. Environmental factors may affect aging - for example, overexposure to ultraviolet radiation accelerates skin aging. Different parts of the body may age at different rates. Two organisms of the same species can also age at different rates, making biological aging and chronological aging distinct concepts.
The occurrence of aging in nature poses an evolutionary puzzle: why would such a deleterious, maladaptive process evolve? This puzzle is deepened by the fact that aging is apparently neither inevitable nor universal: germ lines and several organisms do not exhibit senescent decline.
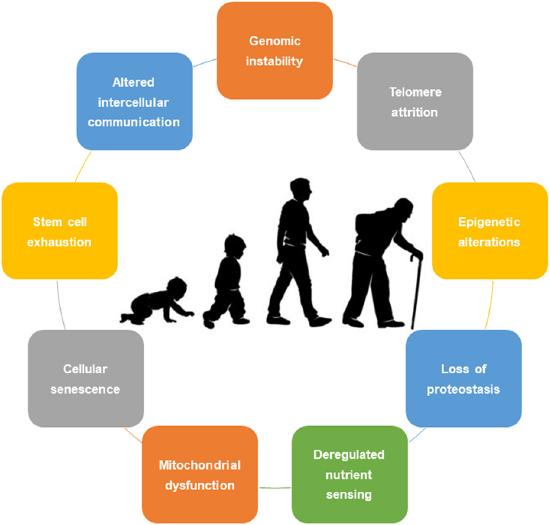
Figure \(\PageIndex{1}\): The nine hallmarks of aging that are common among organisms: genomic instability, telomere attrition, epigenetic alterations, loss of proteostasis, deregulated nutrient sensing, mitochondrial dysfunction, cellular senescence, stem cell exhaustion, and altered intercellular communication. Image: Rebelo-Marques, De Sousa Lages, Andrade, Ribeiro, Mota-Pinto, Carrilho and Espregueira-Mendes - https://www.frontiersin.org/articles/10.3389/fendo.2018.00258/full
Is aging universal? Diverse patterns of senescence among species
Classic theories of aging pertain mainly to relatively short-lived species with increasing mortality and decreasing fertility after maturity, but patterns of aging—including reproductive senescence—are very diverse (Finch 1990, Shefferson et al. 2017, Comfort 1956, Jones et al. 2014, Baudisch et al. 2013, Garcia et al. 2011, Schaible et al. 2015, Ruby et al. 2018, Lemaitre & Gaillard 2017). In particular, although many species do age, some appear to show ‘negligible’ senescence (i.e., only weak or no signs of aging with advancing age; Finch 1990, Shefferson et al. 2017, Finch & Austad 2001, Finch 2009, Finch 1998), whereas others could—at least theoretically—exhibit ‘negative’ senescence (i.e., physiological improvement with age; Vaupel et al. 2004). In freshwater polyps of the genus Hydra (Figure 3.2), for instance, survival and fertility do not decline with age (Schaible et al. 2015). Similarly, many plants (e.g., ~ 93% of angiosperms) show no signs of aging (Salguero-Gómez et al. 2013, Baudisch et al. 2013); some trees, for example, live thousands of years (Figure 3.2). However, a caveat is that aging might in many cases exist but not be detectable because the studied individuals were not old enough (Peron et al. 2010); for example, a recent study of turtles—typically thought of as exhibiting strongly ‘negligible’ senescence—has shown that reproduction and survival do in fact decline with age, contrary to previous expectations. Many organisms, such as numerous invertebrates and fish, start to reproduce before they are fully grown. Increasing body size can then lead to increased fecundity and also to protection against size-specific predators and other sources of mortality. Under these circumstances, the force of natural selection can increase over part of adult life, because the reproductive value of the organism increases (Baudisch 2008, Baudisch 2005, Charlesworth 1994, Partridge & Barton 1996). Non- or slow-aging species, including some animals (e.g., basal metazoans such as Hydra and sea anemones) and most higher plants, are characterized by modular organization, indeterminate (including clonal) growth, and the capacity to regenerate due to stem cell activity; often such organisms start to reproduce before they have finished growing, or they can grow indefinitely (Munné-Bosch 2015, Petralia et al. 2014, Bythell et al. 2017). Some clones of grasses, for example, have been estimated to become 15,000 years old (Noodén 1988). In addition, unlike the standard laboratory model organisms, which set aside and sequestrate their germline early in development, in organisms such as Hydra and higher plants the cells that will become the germline are only identified during adulthood, and these organisms therefore maintain cell lineages with high regenerative potential. Thus, the force of natural selection does not always decline monotonically with age (Baudisch 2008, Munné-Bosch 2015, Shefferson et al. 2017 Baudisch 2005, Charlesworth 1994).




Figure \(\PageIndex{2}\): Long-lived organisms; many organisms age very slowly, if at all. Left: the freshwater polyp Hydra is potentially immortal ("Hydra" by Frank Fox is licensed under CC BY-SA 3.0). Second from left: some trees like this bristlecone pine (Pinus longaeva) live for thousands of years ("Gnarly" by Rick Goldwaser is licensed under CC BY 2.0). Third from left: in the naked mole-rat (Heterocephalus glaber) mortality does not increase with age ("Nacktmull" by Roman Klementschitz is licensed under CC BY-SA 3.0). Right: the bowhead whale (Balaena mysticetus) is the longest-lived mammal, with an estimated maximum life span of 211 years ("A bowhead whale" by Olga Shpak is licensed under CC BY-SA 3.0).
Hypotheses of aging
More than 300 different hypotheses have been posited to explain the nature and causes of aging. Aging hypotheses fall into two broad categories, evolutionary hypotheses of aging and mechanistic hypotheses of aging. Evolutionary hypotheses of aging primarily explain why aging happens, but do not concern themselves with how (the molecular mechanism(s)). All evolutionary hypotheses of aging rest on the basic mechanisms that the force of natural selection declines with age. Mechanistic hypotheses of aging can be divided into hypotheses that propose aging is programmed (i.e., aging follows a biological timetable), and damage accumulation hypotheses (i.e., those that propose aging to be caused by specific molecular changes occurring over time).
Evolutionary aging hypotheses
Antagonistic pleiotropy
One hypothesis proposed by George C. Williams involves antagonistic pleiotropy. Pleiotropy occurs when a single gene has two or more apparently unrelated effects. The idea of antagonistic pleiotropy is that one gene can positively affect a fitness related trait early in life, but can also have negative effects later in life and thus contribute to senescence. Because many more individuals are alive at young ages than at old ages, even small positive effects early can be strongly selected for, and large negative effects later may be very weakly selected against. Williams suggested the following example: Perhaps a gene codes for calcium deposition in bones, which promotes juvenile survival and will therefore be favored by natural selection; however, this same gene promotes calcium deposition in the arteries, causing negative atherosclerotic effects in old age. Thus, harmful biological changes in old age may result from selection for pleiotropic genes that are beneficial early in life but harmful later on. In this case, selection pressure is relatively high when Fisher's reproductive value is high and relatively low when Fisher's reproductive value is low.
Cancer versus cellular senescence trade-off
Senescent cells within a multicellular organism can be purged by competition between cells, but this increases the risk of cancer. This leads to an inescapable dilemma between two possibilities—the accumulation of physiologically useless senescent cells, and cancer—both of which lead to increasing rates of mortality with age.
Disposable soma
The disposable soma hypothesis of aging was proposed by Thomas Kirkwood in 1977. The hypothesis suggests that aging occurs due to a strategy in which an individual only invests in maintenance of the soma for as long as it has a realistic chance of survival. A species that uses resources more efficiently will live longer, and therefore be able to pass on genetic information to the next generation. The demands of reproduction are high, so less effort is invested in repair and maintenance of somatic cells, compared to germline cells, in order to focus on reproduction and species survival.
Damage accumulation hypotheses
The free radical hypothesis
One of the most prominent hypotheses of aging was first proposed by Harman in 1956. It posits that free radicals produced by dissolved oxygen, radiation, cellular respiration and other sources cause damage to the molecular machines in the cell and gradually wear them down. This is also known as oxidative stress. Under normal aerobic conditions, approximately 4% of the oxygen metabolized by mitochondria is converted to superoxide ion, which can subsequently be converted to hydrogen peroxide, hydroxyl radical and eventually other reactive species including other peroxides and singlet oxygen, which can, in turn, generate free radicals capable of damaging structural proteins and DNA.
There is substantial evidence to back up this theory. Old animals have larger amounts of oxidized proteins, DNA and lipids than their younger counterparts.
Chemical damage
One of the earliest aging hypotheses was the Rate of Living Hypothesis described by Raymond Pearl in 1928, which states that fast basal metabolic rate corresponds to shortened maximum life span.
While there may be some validity to the idea that for various types of specific damage detailed below that are byproducts of metabolism, all other things being equal, a fast metabolism may reduce life span, in general this hypothesis does not adequately explain the differences in life span either within, or between, species. Calorically restricted animals process as much, or more, calories per gram of body mass, as their ad libitum fed counterparts, yet exhibit substantially longer life spans. Similarly, metabolic rate is a poor predictor of life span for birds, bats and other species that, it is presumed, have reduced mortality from predation, and therefore have evolved long life spans even in the presence of very high metabolic rates. In a 2007 analysis it was shown that, when modern statistical methods for correcting for the effects of body size and phylogeny are employed, metabolic rate does not correlate with longevity in mammals or birds.
With respect to specific types of chemical damage caused by metabolism, it is suggested that damage to structural proteins or DNA caused by ubiquitous chemical agents in the body such as oxygen and sugars, are in part responsible for aging. The damage can include breakage of biopolymer chains, cross-linking of biopolymers, or chemical attachment of unnatural substituents to biopolymers. Sugars such as glucose and fructose can react with certain amino acids such as lysine and arginine and certain DNA bases such as guanine to produce sugar adducts, in a process called glycation. These adducts can further rearrange to form reactive species, which can then cross-link the structural proteins or DNA to similar biopolymers or other biomolecules such as non-structural proteins. There is evidence that sugar damage is linked to oxidant damage in a process termed glycoxidation.
Mutation accumulation
Natural selection can support lethal and harmful alleles, if their effects are felt after reproduction. The geneticist J. B. S. Haldane wondered why the dominant mutation that causes Huntington's disease remained in the population, and why natural selection had not eliminated it. The onset of this neurological disease is (on average) at age 45 and is invariably fatal within 10–20 years. Haldane assumed that, in human prehistory, few survived until age 45. Since few were alive at older ages and their contribution to the next generation was therefore small relative to the large cohorts of younger age groups, the force of selection against such late-acting deleterious mutations was correspondingly small. Therefore, a genetic load of late-acting deleterious mutations could be substantial at mutation–selection balance. This concept came to be known as the selection shadow (Figure 3.3).
Peter Medawar formalized this observation in his mutation accumulation hypothesis of aging. "The force of natural selection weakens with increasing age—even in a theoretically immortal population, provided only that it is exposed to real hazards of mortality. If a genetic disaster... happens late enough in individual life, its consequences may be completely unimportant".
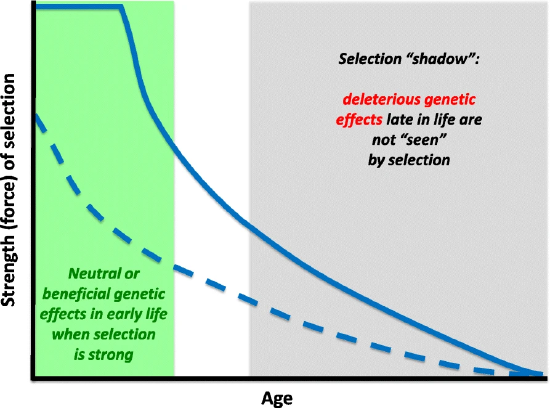
Figure \(\PageIndex{3}\): The declining force of selection. The strength (‘force’) of selection measures how strongly natural selection acts on changes in survival and/or fecundity. Often, but not always, the force of selection declines with age. If this is the case, then alleles with neutral effects on fitness early in life but with deleterious effects late in life can accumulate in a population, unchecked by selection (mutation accumulation). Similarly, alleles with positive effects on fitness components early in life can be selectively favored even if they have negative effects late in life (antagonistic pleiotropy). The late-life negative effects in the ‘selection shadow’ cannot be effectively eliminated by selection, leading to senescence. While the force acting on survival (solid line) only starts to decrease with age after the onset of reproduction, the strength of selection on fecundity (dashed line) can increase or decrease before the onset of reproduction. Image: Flatt and Partridge, https://doi.org/10.1186/s12915-018-0562-z
Trade-offs with life span are pervasive but can be uncoupled
Studies of natural populations have also found support for phenotypic trade-offs consistent with the notion of antagonistic pleiotropy / disposable soma (Nussey et al. 2013, Peron et al. 2010). In bats, for example, species that produce more offspring are shorter-lived than those that produce fewer offspring (Kim et al. 2011). Similarly, a recent review of 26 studies of free-ranging populations of 24 vertebrate species (birds, mammals, reptiles) has identified clear-cut trade-offs between early and late fitness components (Lemaitre et al. 2015), and data in humans have unraveled a genetically based trade-off between reproduction and life span (Wang et al. 2013). Trade-offs thus seem to be pervasive: high resource allocation to growth or reproduction early in life is often associated with earlier or more rapid aging. However, there is also growing evidence that trade-offs between life span and other fitness components are context-dependent and can be ‘uncoupled’, as is observed in some long-lived C. elegans or Drosophila mutants (Flatt & Schmidt 2009, Flatt 2011, Rodrigues & Flatt 2016, Flatt & Heyland 2011), or upon manipulation of specific dietary amino acids in flies (Grandison et al. 2009, Selman et al. 2008; see below), without any apparent fitness costs of longevity. In these cases, a likely explanation is the artificially benign laboratory environment occupied by these organisms, which may allow them to realize their physiologically maximal possible investments into both survival and reproduction.
The most famous example of an ‘uncoupling’ of the fecundity–longevity trade-off is seen in eusocial insects (i.e., ants, bees, termites). In many ants, for example, queens are extraordinarily long-lived and highly fertile as compared to the short-lived and sterile workers (Rodrigues & Flatt 2016, Keller & Genoud 1997, Keller & Jemielity 2006, Kuhn & Korb 2016, Heinze & Schrempf 2008, Schrempf et al. 2017, von Wyschetzki et al. 2015, Hartmann & Heinze 2003, Kramer et al. 2015), even though within the worker caste reproductive costs have been found among fertile bumblebee workers (Blacher et al. 2017). On the other hand, in naked mole rats, which are also eusocial, queens and workers have approximately equivalent life spans but workers do not reproduce while queens can produce up to 900 pups (Buffenstein & Jarvis 2002). How can social insect queens (or kings in termites) escape this trade-off? Surprisingly little formal analysis of this problem exists; the standard explanation that has been put forward is that queens and kings live much longer because they are shielded from extrinsic mortality by the workers (Keller & Genoud 1997, Heinze & Schrempf 2008). In addition, queens or kings may defy the fecundity–longevity trade-off because of trade-offs at the colony level (Kramer et al. 2016), with resources provided by workers freeing them from individual-level trade-offs; at the colony level, queens and kings might be viewed, metaphorically, as representing the ‘immortal germline’, whereas workers can be seen as representing the ‘disposable soma’ (Kramer & Schaible 2013). Classic theories of aging may also not fully apply to eusocial insects (Kramer et al. 2016): their populations exhibit not only age structure but also strong social structure and division of labor. Since in such a situation survival is not only age- but also state-dependent, the force of selection does not necessarily decline with age (Williams & Day 2003). More theoretical work on aging in eusocial insects is warranted, especially the development of class-structured inclusive fitness (kin selection) models (Rodrigues & Flatt 2016, Kramer et al. 2016, Kramer & Schaible 2013, Bourke 2007).
An important insight into the likely explanation for the ‘breaking’ or ‘uncoupling’ of trade-offs comes from the different outcomes of attempts to measure reproductive costs by looking at natural correlations across individuals as opposed to experimental manipulation of reproductive rate. Generally, across individuals in natural populations, there is a positive phenotypic correlation between fecundity and life span. However, the causal connection between the two traits may be the opposite, as experimental manipulations of, for instance, increasing clutch size in birds, often lead to reduced future fecundity or survival (Partridge 1992). This difference occurs because the individual variation in condition and circumstances may obscure the underlying cost of reproduction: healthy individuals in a rich environment may have high fecundity and life span despite the cost of reproduction, which is only revealed by experimental manipulations. This concept has been termed the 'big house, big car effect' (van Noordwijk & de Jong 1986). This underlying cost of reproduction may then constrain the combinations of life history traits that can evolve (van Noordwijk & de Jong 1986, Metcalf 2016). Organisms that live in an environment that is beneficial for development may indeed not experience costs of reproduction (van Noordwijk & de Jong 1986, Metcalf 2016), as often seems to be the case in laboratory animals (Klepsatel et al. 2013). In addition, positive correlations between fitness-related traits can also be caused by mutational variation in recessive deleterious effects (Charlesworth 1990). This arises because such deleterious mutations can have negative pleiotropic effects on two or more traits but the extent of these negative effects varies genetically among individuals.
The big house, big car effect says that individuals differ in the amount of resources (energy, time, or space) they can allocate to competing demands. For example, in humans, each of us has a limited amount of monetary resources. If we were forced to choose between spending our money on a nice house or a nice car (an allocation trade-off), most of us would end up with either a nice house or a nice car, but some people are super wealthy and can afford both a big house and a nice car. If we are only looking across individuals and not accounting for the amount of resources each has accumulated, we often see patterns like this, which suggest an uncoupling of trade-offs. But because individuals differ in their available resources, this observation is not an accurate representation of the true trade-off that is being experience by each individual at the within-individual level.
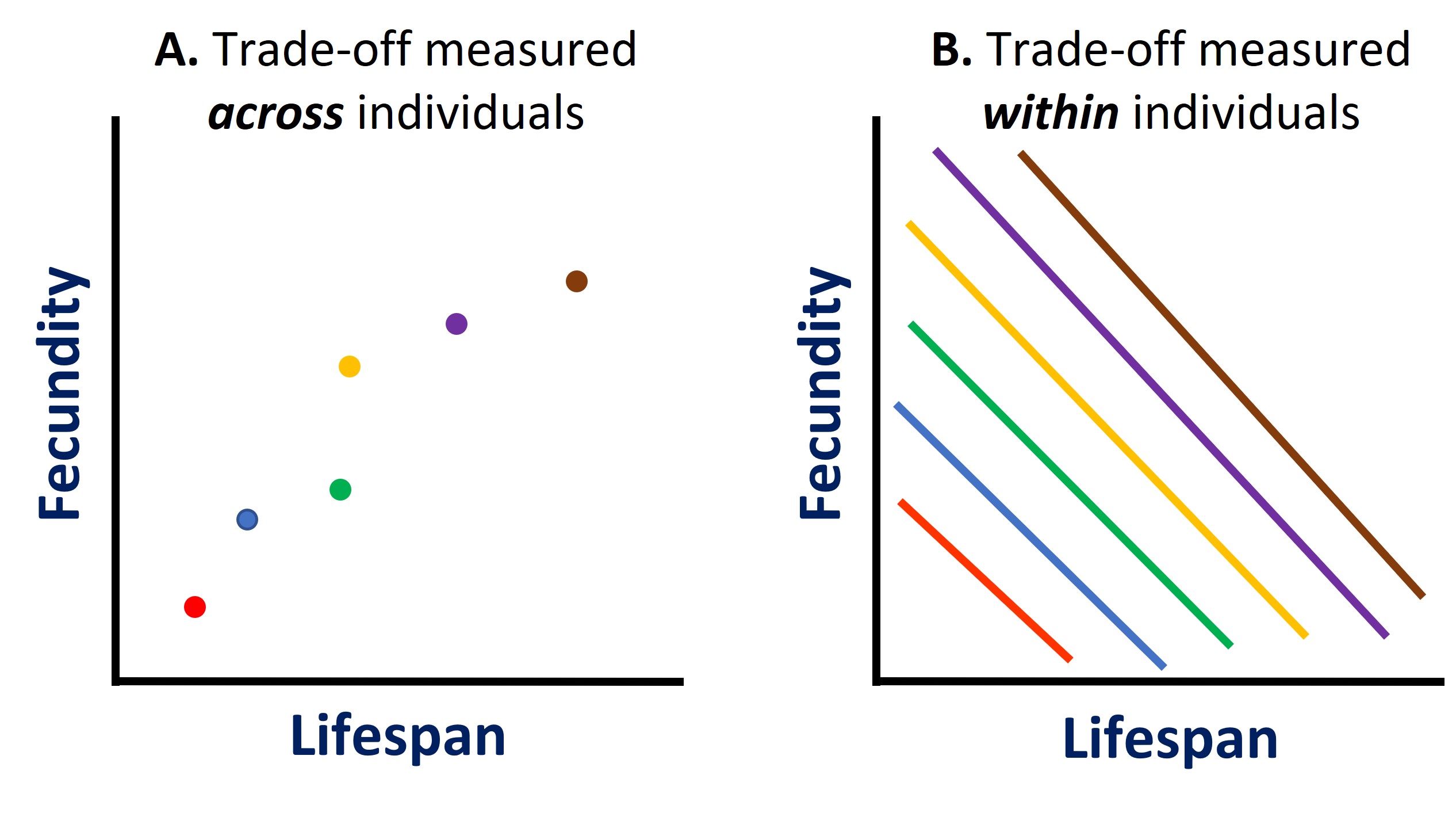
Figure \(\PageIndex{4}\): The 'big house, big car effect' states that individuals differ in the amount of resources they can allocate to competing demands, and thus if we were to correlate life history traits across individuals (A), we can observe an uncoupling of the trade-off. However, because individuals differ in their available resources, this observation is not an accurate representation of the true trade-off that is being experience by each individual at the within-individual level (B). Each color of dot or line represents a different individual with a different amount of resources. Image: Dan Wetzel
The evolution of aging in humans
Human life expectancy worldwide has increased dramatically. During the ~300,000 generations since the divergence from our most recent common ancestor with the great apes, life span evolved to double its previous value (Finch 2010). In the last ~200 years there has been a further substantial increase, on average about 2.5 years per decade, attributable to environmental changes, including improved food, water, hygiene, and living conditions, reduced impact of infectious disease with immunization and antibiotics, and improved medical care at all ages (Vaupel et al. 1998, Wilmoth 2000, Oeppen & Vaupel 2002, Vaupel 2010). Many modern humans inhabit a very different environment from that in which their life history evolved, with both protection from many of its dangers, such as predators, infectious diseases, and harsh physical conditions, and freedom from the need to forage extensively to avoid starvation (Finch 2010). As a result, most people are now living long beyond the ages at which most would have been dead in the past. Natural selection has therefore not had an opportunity to maintain evolutionary fitness at older ages.
In humans, where age-related changes are particularly well documented, aging has proved to be a complex process of functional decline and accumulation of diverse pathologies in different tissues (Lopez-Otin et al. 2013, Kirkwood et al. 1999). Williams predicted in 1957 that aging is likely to be a genetically complex trait, and different lineages and taxa might well exhibit different proximate mechanisms of senescence. Indeed, natural variation in the rate of aging is likely influenced by many genes (Burke et al. 2014, Highfill et al. 2016, Ivanov et al. 2015), since survival and reproduction between them harness the activity of much of the genome.


Figure \(\PageIndex{5}\): Aging in humans. Images: RODNAE Productions and Andrea Piacquadio.
Prevention of late-life morbidity in humans ideally would involve interventions that could be started at the earliest in middle age. Pharmacological prevention of cardiovascular disease, with statins and blood pressure lowerers, is already routine in clinical practice (Sundstrom et al. 2018). Unsurprisingly, many of the proteins that have turned out to be important in aging also play prominent roles in the etiology of age-related diseases, and are already the targets of licensed drugs. Consideration is hence starting to be given to widening the preventative, pharmacological approach, for instance by repurposing drugs that are used to treat cancer, prevent rejection of transplanted organs, and diabetes because they have been found to extend life span in model organisms (Blenis 2017, Mannick et al. 2014, Barzilai et al. 2016, Johnson & Kaeberlein 2016). Other possible approaches to emerge from experimental work with animals include removal of damaging senescent cells that accumulate during aging (Childs et al. 2015, Rando & Chang 2012), use of factors from young blood that restore the age-related loss of function of stem cells or synapses between nerve cells in the brain (Rando & Chang 2012, Mair & Dillin 2008), and alteration of the composition of the microorganisms in the gut to a younger profile (Clark & Walker 2018, Kundu et al. 2017, Schmidt et al. 2018), which has already been shown to extend life span in the turquoise killifish (Smith et al. 2017).
However, despite the considerable promise of these approaches, the extent to which they can yield health benefits free of side effects needs detailed study, since they could pose some new challenges for an aged system. For instance, removal of senescent cells, or restoration of stem cell function, could be beneficial in the short term, but in the longer term could lead to stem cell exhaustion and tissue dysfunction.
Sources
-
Barzilai, N., Crandall, J.P., Espeland, M.A., & Kritchevsky, S.B. (2016). Metformin as a tool to target aging. Cell Metab, 23(6), pp. 1060–5.
- Baudisch A. (2005). Hamilton’s indicators of the force of selection. Proc Natl Acad Sci USA, 102(23), pp. 8263–8.
- Baudisch A. (2008). Inevitable aging? Berlin: Springer.
-
Baudisch, A., Salguero-Gómez, R., Jones, O.R., Wrycza, T., Mbeau-Ache, C., Franco, M., et al. (2013). The pace and shape of senescence in angiosperms. J Ecol, 101(3), pp. 596–06.
-
Blacher, P., Bourke, A.F.G, & Huggins, T.J. (2017). Evolution of ageing, costs of reproduction and the fecundity–longevity trade-off in eusocial insects. Proc R Soc Lond B, 284. 20170380.
-
Blenis, J. (2017). The gateway to cellular metabolism, cell growth, and disease. Cell, 171(1), pp. 10–3.
-
Bourke, A.F.G. (2007). Kin selection and the evolutionary theory of aging. Annu Rev Ecol Evol Syst, 38, pp. 103–28.
-
Buffenstein, R., & Jarvis, J.U.M. (2002). The naked mole rat: A new record for the oldest living rodent. Sci Aging Knowl Environ, 2002(21):pe7.
-
Burke, M.K., King, E.G., Long, A.D., Rose, M.R., & Shahrestani, P. (2014). Genome-wide association study of extreme longevity in Drosophila melanogaster. Genome Biol Evol., 6(1), pp. 1–11.
-
Bythell, J.C., Brown, B.E., & Kirkwood, T.B.L. (2017). Do reef corals age? Biol Rev., 93(2), pp. 1192–02.
-
Charlesworth, B. (1990). Optimization models, quantitative genetics, and mutation. Evolution, 44, pp. 520–38.
-
Charlesworth, B. (1994). Evolution in age-structured populations. 2nd ed. Cambridge: Cambridge University Press.
-
Childs, B.G., Durik, M., Baker, D.J., & van Deursen, J.M. (2015). Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat Med, 21, pp. 1424–35.
-
Childs, B.G., Gluscevic, M., Baker, D.J., Laberge, R.M., Marquess, D., Dananberg, J., & van Deursen, J.M. (2017). Senescent cells: an emerging target for diseases of ageing. Nat Rev Drug Discov., 16, pp. 718–35.
-
Clark, R.I., & Walker, D.W. (2018). Role of gut microbiota in aging-related health decline: insights from invertebrate models. Cell Mol Life Sci., 75(1), pp. 93–101.
-
Comfort, A. (1956). The biology of senescence. London: Routledhe & Kegan Paul.
-
Finch, C.E. (2010). Evolution of the human life span and diseases of aging: roles of infection, inflammation, and nutrition. Proc Natl Acad Sci USA, 107(suppl 1), pp. 1718–24.
-
Finch, C.E. (1990). Longevity, senescence, and the genome. Chicago and London: The University of Chicago Press.
-
Finch, C.E. (2009). Update on slow aging and negligible senescence: A mini-review. Gerontology, 55(3), pp. 307–13.
-
Finch, C.E. (1998). Variations in senescence and longevity include the possibility of negligible senescence. J Gerontol A Biol Sci Med Sci., 53(4):B235–9.
-
Finch, C.E., & Austad, S.N. (2001). History and prospects: Symposium on organisms with slow aging. Exp Gerontol, 36(4), pp. 593–7.
-
Flatt, T. (2011). Survival costs of reproduction in Drosophila. Exp Gerontol, 46(5), pp. 369–75.
- Flatt, T., & Heyland, A. (2011). Mechanisms of life history evolution: The genetics and physiology of life history traits and trade-offs. Oxford: Oxford University Press.
-
Flatt, T., & Schmidt, P.S. (2009). Integrating evolutionary and molecular genetics of aging. Biochim Biophys Acta., 1790(10), pp. 951–62.
- Dahlgren, J.P., Ehrlén, J., & Garcia, M.B. (2011). No evidence of senescence in a 300-year-old mountain herb. J Ecol., 99(6), pp. 1424–30.
-
Grandison, R.C., Partridge, L., & Piper, M.D.W. (2009). Amino-acid imbalance explains extension of life span by dietary restriction in Drosophila. Nature, 462(7276), pp. 1061–5.
-
Hartmann. A., & Heinze, J. (2003). Lay eggs, live longer: division of labor and life span in a clonal ant species. Evolution, 57, pp. 2424–9.
-
Heinze, J., & Schrempf, A. (2008). Aging and reproduction in social insects: A mini-review. Gerontology, 54, pp. 160–7.
-
Highfill, C.A., Macdonald, S.J., & Reeves, G.A. (2016). Genetic analysis of variation in life span using a multiparental advanced intercross Drosophila mapping population. BMC Genet., 17(1):113.
-
Ivanov, D.K., Escott-Price, V., Ziehm, M., Magwire, M.M., Mackay, T.F.C., Partridge, L., et al. (2015). Longevity GWAS using the Drosophila genetic reference panel. J Gerontol A Biol Sci Med Sci., 70(12), pp. 1470–8.
-
Johnson, S.C., & Kaeberlein, M. (2016). Rapamycin in aging and disease: Maximizing efficacy while minimizing side effects. Oncotarget, 7(29), pp. 44876–8.
-
Jones, O.R., Scheuerlein, A., Salguero-Gomez, R., Camarda, C.G., Schaible, R., Casper, B.B., et al. (2014). Diversity of ageing across the tree of life. Nature, 505(7482), pp. 169–73.
-
Keller, L., & Genoud, M. (1997). Extraordinary life spans in ants: A test of evolutionary theories of ageing. Nature, 389, pp. 958–60.
-
Keller, L., & Jemielity, S. (2006). Social insects as a model to study the molecular basis of ageing. Exp Gerontol, 41, pp. 553–6.
-
Kirkwood, T.B.L., Martin, G.M., & Partridge, L. (1999). Evolution, senescence and health in old age. In: Stearns SC, editor. Evolution in health and disease. Oxford: Oxford University Press, p. 219–30.
-
Klepsatel, P., Gáliková, M., Maio, N., Schlötterer, C., & Flatt, T. (2013). Reproductive and post-reproductive life history of wild-caught Drosophila melanogaster under laboratory conditions. J Evol Biol., 26(7), pp. 1508–20.
-
Kramer, B.H., & Schaible, R. (2013). Life span evolution in eusocial workers: A theoretical approach to understanding the effects of extrinsic mortality in a hierarchical system. PLoS One, 8:e61813.
-
Kramer, B.H., Schrempf, A., Scheuerlein, A., & Heinze, J.(2015). Ant colonies do not trade-off reproduction against maintenance. PLoS One, 10:e0137969.
-
Kramer, B.H., van Doorn, G.S., & Weissing, F.J. (2016). Pen I. life span divergence between social insect castes: challenges and opportunities for evolutionary theories of aging. Curr Opin Insect Sci., 16, pp. 76–80.
-
Kuhn, J.M.M., & Korb, J. (2016). Editorial overview: social insects: aging and the re-shaping of the fecundity/longevity trade-off with sociality. Curr Opin Insect Sci., 16, pp. vii–x.
-
Kundu, P., Blacher, E., Elinav, E., & Pettersson, S. (2017). Our gut microbiome: The evolving inner self. Cell, 171(7), pp. 1481–93.
-
Lemaitre, J.F., Berger, V., Bonenfant, C., Douhard, M., Gamelon, M., Plard, F. et al. (2015). Early-late life trade-offs and the evolution of ageing in the wild. Proc R Soc Lond B., 282(1806):20150209.
-
Lemaître, J.F., & Gaillard, J.M. (2017). Reproductive senescence: new perspectives in the wild. Biol Rev., 92, pp. 2182–99.
-
Lopez-Otin, C., Blasco, M.A., Partridge, L., Serrano, M., & Kroemer, G. (2013). The hallmarks of aging. Cell, 153(6), pp. 1194–217.
-
Mannick, J.B., Del Giudice, G., Lattanzi, M., Valiante, N.M., Praestgaard, J., Huang, B., Lonetto, M.A., Maecker, H.T., Kovarik, J., Carson, S., et al. (2014). mTOR inhibition improves immune function in the elderly. Sci Transl Med., 6(268):268ra179.
-
Metcalf, C.J.E. (2016). Invisible trade-offs: Van Noordwijk and de Jong and life-history evolution. Am Nat., 187(4):iii–v.
- Munné-Bosch, S. (2015). Senescence: Is it universal or not? Trends Plant Sci., 20(11), pp. 713–20.
-
Noodén, L.D. (1988). Whole plant senescence. In Thiman, K.V., & Leopold, A.C. (Eds.), Senescence and aging in plants. Cambridge: Academic Press, p. 391–439.
-
Nussey, D.H., Froy, H., Lemaitre, J.F., Gaillard, J.M., & Austad, S.N. (2013). Senescence in natural populations of animals: Widespread evidence and its implications for bio-gerontology. Ageing Res Rev., 12(1), pp. 214–25.
-
Oeppen, J., Vaupel, J.W. (2002). Broken limits to life expectancy. Science, 296(5570), pp. 1029–31.
-
Partridge, L. (1992). Measuring reproductive costs. Trends Ecol Evol., 7(3), pp. 99–100.
-
Partridge, L., & Barton, N.H. (1996). On measuring the rate of ageing. Proc R Soc Lond B., 263(1375), pp. 1365–71.
-
Peron, G., Gimenez, O., Charmantier, A., Gaillard, J.M., & Crochet, P.A. (2010). Age at the onset of senescence in birds and mammals is predicted by early-life performance. Proc R Soc Lond B., 277, pp. 2849–56.
-
Petralia, R.S., Mattson, M.P., & Yao, P.J. (2014). Aging and longevity in the simplest animals and the quest for immortality. Ageing Res Rev., 16, pp. 66–82.
-
Rodrigues, M.A., & Flatt, T. (2016). Endocrine uncoupling of the trade-off between reproduction and somatic maintenance in eusocial insects. Curr Opin Insect Sci., 16, pp. 1–8.
-
Ruby, J.G., Smith, M., & Buffenstein, R. (2018). Naked mole-rat mortality rates defy Gompertzian laws by not increasing with age. elife., 7:e31157.
-
Salguero-Gómez, R., Shefferson, R.P., & Hutchings, M.J. (2013). Plants do not count … or do they? New perspectives on the universality of senescence. J Ecol., 101(3), pp. 545–54.
-
Schaible, R., Scheuerlein, A., Dańko, M.J., Gampe, J., Martínez, D.E., & Vaupel, J.W. (2015). Constant mortality and fertility over age in Hydra. Proc Natl Acad Sci USA, 112(51), pp. 15701–6.
-
Schmidt, T.S.B., Raes, J., & Bork, P. (2018). The human gut microbiome: From association to modulation. Cell, 172(6), pp. 1198–215.
-
Schrempf, A., Giehr, J., Röhrl, R., Steigleder, S., & Heinze, J. (2017). Royal Darwinian demons: enforced changes in reproductive efforts do not affect the life expectancy of ant queens. Am Nat., 189(4), pp. 436–42.
-
Selman, C., Lingard, S., Choudhury, A.I., Batterham, R.L., Claret, M., Clements, M., et al. (2008). Evidence for life span extension and delayed age-related biomarkers in insulin receptor substrate 1 null mice. FASEB J., 22(3), pp. 807–18.
-
Shefferson, R.P., Jones, O.R., & Salguero-Gómez, R. (2017). The evolution of senescence in the tree of life. Cambridge: Cambridge University Press.
-
Smith, P., Willemsen, D., Popkes, M., Metge, F., Gandiwa, E., Reichard, M., & Valenzano, D.R. (2017). Regulation of life span by the gut microbiota in the short-lived African turquoise killifish. elife., 6:e27014.
-
Sundström, J., Gulliksson, G., & Wirén, M. (2018). Synergistic effects of blood pressure-lowering drugs and statins: Systematic review and meta-analysis. BMJ Evid Based Med., 23(2), pp. 64–9.
-
Thomas, Chang. (2012). Aging, rejuvenation, and epigenetic reprogramming: Resetting the aging clock. Cell, 148(1), pp. 46–57.
-
Van Noordwijk, A., & de Jong, G. (1986). Acquisition and allocation of resources: their influence on variation in life history tactics. Am Nat., 128(1), pp. 137–42.
-
Vaupel, J.W. (2010). Biodemography of human ageing. Nature, 464(7288), pp. 536–42.
-
Vaupel, J.W., Baudisch, A., Dolling, M., Roach, D.A., & Gampe, J. (2004). The case for negative senescence. Theor Popul Biol., 65(4), pp. 339–51.
-
Vaupel, J.W., Carey, J.R., Christensen, K., Johnson, T.E., Yashin, A.I., Holm, N.V., et al. (1998). Biodemographic trajectories of longevity. Science, 280(5365), pp. 855–60.
- Von Wyschetzki, K., Rueppell, O., Oettler, J., & Heinze, J. (2015). Transcriptomic signatures mirror the lack of the fecundity/longevity trade-off in ant queens. Mol Biol Evol., 32, pp. 3173–85.
-
Wang, X., Byars, S.G., & Stearns, S.C. (2013). Genetic links between post-reproductive life span and family size in Framingham. Evol Med Pub Health, (1), pp. 241–53.
-
Warner, D.A., Miller, D.A.W., Bronikowski, A.M., & Janzen, F.J. (2016). Decades of field data reveal that turtles senesce in the wild. Proc Natl Acad Sci USA, 113, pp. 6502–7.
-
Williams, G.C. (1957). Pleiotropy, natural selection, and the evolution of senescence. Evolution, 11, pp. 398–411.
-
Williams, P.D., & Day, T. (2003). Antagonistic pleiotropy, mortality source interactions, and the evolutionary theory of senescence. Evolution, 57(7), pp. 1478–88.
-
Wilkinson, G.S., & South, J.M. (2002). Life history, ecology and longevity in bats. Aging Cell, 1(2), pp. 124–31.
-
Wilmoth, J.R. (2000). Demography of longevity: past, present, and future trends. Exp Gerontol., 35(9-10), pp. 1111–29.
Contributors and Attributions
This chapter was written by D. Wetzel with text taken from the following CC-BY resources:
- Flatt, T., Partridge, L. (2018). Horizons in the evolution of aging. BMC Biol, 16(93). https://doi.org/10.1186/s12915-018-0562-z
- Andrade, R., Carrilho, F., De Sousa Lages, A., Espreguiera-Mendes, J., Mota-Pinto, A., Rebelo-Marques, A., & Ribeiro C.F. (2018). Aging hallmarks: The benefits of physical exercise. Front. Endocrinol, 9(258). https://www.frontiersin.org/articles/10.3389/fendo.2018.00258/full
- Wikipedia: https://en.wikipedia.org/wiki/Senescence

