3.2: The Viruses
- Last updated
- Save as PDF
- Page ID
- 102602
Viral Classification
Since viruses lack ribosomes (and thus rRNA), they cannot be classified within the Three Domain Classification scheme with cellular organisms. Alternatively, Dr. David Baltimore derived a viral classification scheme, one that focuses on the relationship between a viral genome to how it produces its mRNA. The Baltimore Scheme recognizes seven classes of viruses.

DNA viruses
Class I: dsDNA
DNA viruses with a dsDNA genome, like bacteriophages T4 and lambda, have a genome exactly the same as the host cell that they are infecting. For this reason, many host enzymes can be utilized for replication and/or protein production. The flow of information follows a conventional pathway: dsDNA → mRNA → protein, with a DNA-dependent RNA-polymerase producing the mRNA and the host ribosome producing the protein. The genome replication, dsDNA → dsDNA, requires a DNA-dependent DNA-polymerase from either the virus or the host cell.

dsDNA.
The virus often employs strategies for control of gene expression, to insure that particular viral products are made at specific times in the virus replication. In the case of T4, the host RNA polymerase binds to the viral DNA and begins transcribing early genes immediately after the DNA is injected into the cell. One of the early viral proteins modifies the host RNA polymerase so that it will no longer recognize host promoters at all, in addition to moving on to transcribe genes for middle-stage viral proteins. A further modification (catalyzed by middle-stage viral proteins) further modified the RNA polymerase so it will recognize viral genes coding for late-stage proteins. This insures an orderly production of viral proteins.
The replication of several dsDNA viruses results in the production of concatemers, where several viral genomes are linked together due to short single-stranded regions with terminal repeats. As the genome is packaged into the capsid a viral endonuclease cuts the concatemer to an appropriate length.
There are several animal viruses with dsDNA genomes, such as the pox viruses and the adenoviruses. The herpesviruses have several notable features, such as the link of several members with cancer and the ability of the viruses to remain in a latent form within their host. A productive infection results in an explosive viral population, cell death, and development of disease signs, during which neurons are infected. A latent infection develops in the neurons, allowing the virus to remain undetected in the host. If the viral genome is reactivated, a productive infection results, leading to viral replication and disease signs again.
Class II: ssDNA
The flow of information for ssDNA viruses, such as the parvoviruses, will still follow the conventional pathway, to a certain extent: DNA → mRNA → protein. But the viral genome can either have the same base sequence as the mRNA (plus-strand DNA) or be complementary to the mRNA (minus-strand DNA). In the former case, a DNA strand that is complementary to the viral genome must be manufactured first, forming a double-stranded replicative form (RF). This can be used to both manufacture viral proteins and as a template for viral genome copies. For the minus-strand DNA viruses, the genome can be used directly to produce mRNA but a complementary copy will still need to be made, to serve as a template for viral genome copies.

ssDNA.
The replicative form can be used for rolling-circle replication, where one strand is nicked and replication enzymes are used to extend the free 3’ end. As a complementary strand is synthesized around the circular DNA, the 5’ end is peeled off, leading to a displaced strand that continues to grow in length.
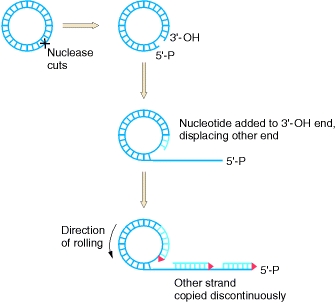
Rolling-Circle Replication.
Class VII: DNA viruses that use reverse transcriptase
The hepadnaviruses contain a DNA genome that is partially double-stranded, but contains a single-stranded region. After gaining entrance into the cell’s nucleus, host cell enzymes are used to fill in the gap with complementary bases to form a dsDNA closed loop. Gene transcription yields a plus-strand RNA known as the pregenome, as well as the viral enzyme reverse transcriptase, an RNA-dependent DNA-polymerase. The pregenome is used as a template for the reverse transcriptase to produced minus-strand DNA genomes, with a small piece of pregenome used as a primer to produce the double-stranded region of the genomes.
RNA viruses
Class III: dsRNA
Double-stranded RNA viruses infect bacteria, fungi, plants, and animals, such as the rotavirus that causes diarrheal illness in humans. But cells do not utilize dsRNA in any of their processes and have systems in place to destroy any dsRNA found in the cell. Thus the viral genome, in its dsRNA form, must be hidden or protected from the cell enzymes. Cells also lack RNA-dependent RNA-polymerases, necessary for replication of the viral genome so the virus must provide this enzyme itself. The viral RNA-dependent RNA polymerase acts as both a transcriptase to transcribe mRNA, as well as a replicase to replicate the RNA genome.

dsRNA.
For the rotavirus, the viral nucleocapsid remains intact in the cytoplasm with replication events occurring inside, allowing the dsRNA to remain protected. Messenger RNA is transcribed from the minus-strand of the RNA genome and then translated by the host ribosome in the cytoplasm. Viral proteins aggregate to form new nucleocapsids around RNA replicase and plus-strand RNA. The minus-strand RNA is then synthesized by the RNA replicase within the nucleocapsid, once again insuring protection of the dsRNA genome.
Class IV: +ssRNA
Viruses with plus-strand RNA, such as poliovirus, can use their genome directly as mRNA with translation by the host ribosome occurring as soon as the unsegmented viral genome gains entry into the cell. One of the viral genes expressed yields an RNA-dependent RNA-polymerase (or RNA replicase), which creates minus-strand RNA from the plus-strand genome. The minus-strand RNA can be used as a template for more plus-strand RNA, which can be used as mRNA or as genomes for the newly forming viruses.

+ssRNA.
Translation of the poliovirus genome yields a polyprotein, a large protein with protease activity that cleaves itself into three smaller proteins. Additional cleavage activity eventually yields all the proteins needed for capsid formation, as well as an RNA-dependent RNA-polymerase.
The formation of a polyprotein that is cut into several smaller proteins illustrates one possible strategy to an issue faced by many +ssRNA viruses – how to generate multiple proteins from an unsegmented +ssRNA genome? Other possibilities include:
- subgenomic mRNA – during translation, portions of the viral RNA may be skipped, resulting in different proteins than what is made from the viral RNA in its entirety.
- ribosomal frame-shifting – the ribosome “reads” the mRNA in groups of three nucleotides or codon, which translate to one amino acid. If the ribosome starts with nucleotide #1, that is one open reading frame (ORF), resulting in one set of amino acids. If the ribosome were to move forward where nucleotide 2 is the starting nucleotide that would be ORF #2, resulting in a completely different set of amino acids. If the ribosome were to move forward again where nucleotide 3 is the starting nucleotide that would be ORF#3, resulting in an entirely different set of amino acids. Some viruses have viral genes that deliberately overlap within different ORFs, leading to the production of different proteins from a single mRNA.
- readthrough mechanism – a viral genome can have stop codons embedded throughout the sequence. When the ribosome comes to a stop codon it can either stop, ending the amino acid sequence, or it can ignore the stop codon, continuing on to make a longer string of amino acids. For viruses with the readthrough mechanism, they acquire a variety of proteins by having stop codons that are periodically ignored. Sometimes this function is combined with the ribosomal frame-shifting to produce an even greater variety of viral proteins.
Class V: -ssRNA
Minus-strand RNA viruses include many members notable for humans, such as influenza virus, rabies virus, and Ebola virus. Since the genome of minus-strand RNA viruses cannot be used directly as mRNA, the virus must carry an RNA-dependent RNA-polymerase within its capsid. Upon entrance into the host cell, the plus-strand RNAs generated by the polymerase are used as mRNA for protein production. When viral genomes are needed the plus-strand RNAs are used as templates to make minus-strand RNA.

-ssRNA.
Class VI: +ssRNA, retroviruses
Despite the fact that the retroviral genome is composed of +ssRNA, it is not used as mRNA. Instead, the virus uses its reverse transcriptase to synthesize a piece of ssDNA complementary to the viral genome. The reverse transcriptase also possesses ribonuclease activity, which is used to degrade the RNA strand of the RNA-DNA hybrid. Lastly, the reverse transcriptase is used as a DNA polymerase to make a complementary copy to the ssDNA, yielding a dsDNA molecule. This allows the virus to insert its genome, in a dsDNA form, into the host chromosome, forming a provirus. Unlike a prophage, a provirus can remain latent indefinitely or cause the expression of viral genes, leading to the production of new viruses. Excision of the provirus does not occur for gene expression.

+ssRNA, retroviruses.
Other Infectious Agents
Viroids
Viroids are small, circular ssRNA molecules that lack protein. These infectious molecules are associated with a number of plant diseases. Since ssRNA is highly susceptible to enzymatic degradation, the viroid RNA has extensive complementary base pairing, causing the viroid to take on a hairpin configuration that is resistant to enzymes. For replication viroids rely on a plant RNA polymerase with RNA replicase activity.
Prions
Prions are infectious agents that completely lack nucleic acid of any kind, being made entirely of protein. They are associated with a variety of diseases, primarily in animals, although a prion has been found that infects yeast (!). Diseases include bovine spongiform encephalopathy (BSE or “mad cow disease”), Creutzfeld-Jakob disease in humans, and scrapie in sheep.
The prion protein is found in the neurons of healthy animals (PrPC or Prion Protein Cellular), with a particular secondary structure. The pathogenic form (PrPSC or Prion Protein Scrapie) has a different secondary structure and is capable of converting the PrPC into the pathogenic form. Accumulation of the pathogenic form causes destruction of brain and nervous tissue, leading to disease symptoms such as memory loss, lack of coordination, and eventually death.

Prions. Joannamasel at English Wikipedia [CC BY-SA 3.0], via Wikimedia Commons
Key Words
Baltimore Scheme, Class I, Class II, Class III, Class IV, Class V, Class VI, Class VII, DNA-dependent RNA polymerase, DNA-dependent DNA-polymerase, concatemer, productive infection, latent infection, plus-strand DNA/+DNA, minus-strand DNA/-DNA, dsDNA, ssDNA, replicative form (RF), rolling-circle replication, pregenome, reverse transcriptase, RNA-dependent DNA-polymerase, dsRNA, RNA-dependent RNA-polymerase, transcriptase, replicase, plus-strand RNA/+ssRNA, minus-strand RNA/-ssRNA, polyprotein, subgenomic mRNA, ribosomal frame-shifting, open reading frame (ORF), readthrough mechanism, stop codon, retrovirus, ribonuclease, provirus, viroid, prion, PrPC/Prion Protein Cellular, PrPSC/Prion Protein Scrapie.
Essential Questions/Objectives
- What is the Baltimore system of classification? What viral characteristics does it use?How does each viral group make proteins and replicate their genome? Where do the necessary components come from? (virus or host cell) What modifications are necessary, for viruses with a genome different from the host cell?
- What strategy do dsDNA viruses use for control of gene expression? What are concatemers? What are productive and latent infections?
- What is a replicative form? What is rolling circle replication? What is the advantage of these viral mechanisms?
- What is a pregenome? What is reverse transcriptase? What role does it play for the Class VII viruses?
- What issues do dsRNA viruses face? How do they overcome these issues? What is a transcriptase? What is a replicase?
- How is the genome used by Class IV +ssRNA viruses? What are the strategies used by these viruses to generate multiple proteins from an unsegmented genome?
- What steps are necessary for the –ssRNA viruses?
- How do the retroviruses, as +ssRNA viruses, differ from the Class IV viruses? What is a ribonuclease? What is a provirus?
- What is a viroid? What is a prion? How do these agents cause disease? How do they replicate?
Exploratory Questions (OPTIONAL)
- Why were scientists initially so resistant to the idea of prions lacking any type of nucleic acid?


