6: Bacteria - Surface Structures
- Page ID
- 10646
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Layers Outside the Cell Wall
What have we learned so far, in terms of cell layers? All cells have a cell membrane. Most bacteria have a cell wall. But there are a couple of additional layers that bacteria may, or may not, have. These would be found outside of both the cell membrane and the cell wall, if present.
Capsule
A bacterial capsule is a polysaccharide layer that completely envelopes the cell. It is well organized and tightly packed, which explains its resistance to staining under the microscope. The capsule offers protection from a variety of different threats to the cell, such as desiccation, hydrophobic toxic materials (i.e. detergents), and bacterial viruses. The capsule can enhance the ability of bacterial pathogens to cause disease and can provide protection from phagocytosis (engulfment by white blood cells known as phagocytes). Lastly, it can help in attachment to surfaces.
Slime Layer
A bacterial slime layer is similar to the capsule in that it is typically composed of polysaccharides and it completely surrounds the cell. It also offers protection from various threats, such as desiccation and antibiotics. It can also allow for adherence to surfaces. So, how does it differ from the capsule? A slime layer is a loose, unorganized layer that is easily stripped from the cell that made it, as opposed to a capsule which integrates firmly around the bacterial cell wall.

S-Layer
Some bacteria have a highly organized layer made of secreted proteins or glycoproteins that self-assemble into a matrix on the outer part of the cell wall. This regularly structured S-layer is anchored into the cell wall, although it is not considered to be officially part of the cell wall in bacteria. S-layers have very important roles for the bacteria that have them, particularly in the areas of growth and survival, and cell integrity.
S layers help maintain overall rigidity of the cell wall and surface layers, as well as cell shape, which are important for reproduction. S layers protect the cell from ion/pH changes, osmotic stress, detrimental enzymes, bacterial viruses, and predator bacteria. They can provide cell adhesion to other cells or surfaces. For pathogenic bacteria they can provide protection from phagocytosis.
Structures Outside the Cell Wall
Bacteria can also have structures outside of the cell wall, often bound to the cell wall and/or cell membrane. The building blocks for these structures are typically made within the cell and then secreted past the cell membrane and cell wall, to be assembled on the outside of the cell.
Fimbriae (sing. fimbria)
Fimbriae are thin filamentous appendages that extend from the cell, often in the tens or hundreds. They are composed of pilin proteins and are used by the cell to attach to surfaces. They can be particularly important for pathogenic bacteria, which use them to attach to host tissues.
Pili (sing. pilin)
Pili are very similar to fimbriae (some textbooks use the terms interchangeably) in that they are thin filamentous appendages that extend from the cell and are made of pilin proteins. Pili can be used for attachment as well, to both surfaces and host cells, such as the Neisseria gonorrhea cells that use their pili to grab onto sperm cells, for passage to the next human host. So, why would some researchers bother differentiating between fimbriae and pili?
Pili are typically longer than fimbriae, with only 1-2 present on each cell, but that hardly seems enough to set the two structures apart. It really boils down to the fact that a few specific pili participate in functions beyond attachment. The conjugative pili participate in the process known as conjugation, which allows for the transfer of a small piece of DNA from a donor cell to a recipient cell. The type IV pili play a role in an unusual type of motility known as twitching motility, where a pilus attaches to a solid surface and then contracts, pulling the bacterium forward in a jerky motion.
Flagella (sing. flagellum)
Bacterial motility is typically provided by structures known as flagella. The bacterial flagellum differs in composition, structure, and function from the eukaryotic flagellum, which operates as a flexible whip-like tail utilizing microtubules. The bacterial flagellum is rigid in nature and operates more like the propeller on a boat.
There are three main components to the bacterial flagellum:
- the filament – a long thin appendage that extends from the cell surface. The filament is composed of the protein flagellin and is hollow. Flagellin proteins are transcribed in the cell cytoplasm and then transported across the cell membrane and cell wall. A bacterial flagellar filament grows from its tip (unlike the hair on your head), adding more and more flagellin units to extend the length until the correct size is reached. The flagellin units are guided into place by a protein cap.
- the hook – this is a curved coupler that attaches the filament to the flagellar motor.
- the motor – a rotary motor that spans both the cell membrane and the cell wall, with additional components for the gram negative outer membrane. The motor has two components: the basal body, which provides the rotation, and the stator, which provides the torque necessary for rotation to occur. The basal body consists of a central shaft surrounded by protein rings, two in the gram positive bacteria and four in the gram negative bacteria. The stator consists of Mot proteins that surround the ring(s) embedded within the cell membrane.
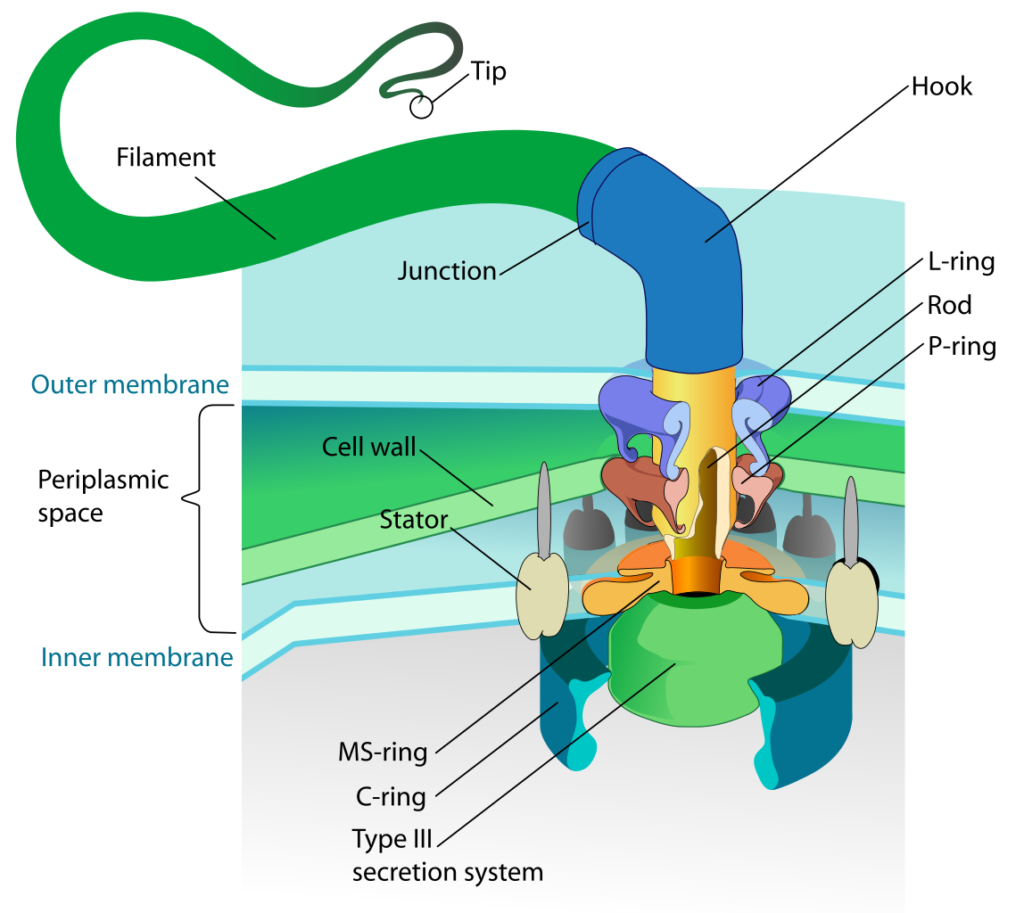
Flagellum base diagram. By LadyofHats (Own work) [Public Domain], Via Wikipedia Commons
Bacterial Movement
Bacterial movement typically involves the use of flagella, although there are a few other possibilities as well (such as the use of type IV pili for twitching motility). But certainly the most common type of bacterial movement is swimming, which is accomplished with the use of a flagellum or flagella.
Swimming
Rotation of the flagellar basal body occurs due to the proton motive force, where protons that accumulate on the outside of the cell membrane are driven through pores in the Mot proteins, interacting with charges in the ring proteins as they pass across the membrane. The interaction causes the basal body to rotate and turns the filament extending from the cell. Rotation can occur at 200-1000 rpm and result in speeds of 60 cell lengths/second (for comparison, a cheetah moves at a maximum rate of 25 body lengths/second).
Rotation can occur in a clockwise (CW) or a counterclockwise (CCW)direction, with different results to the cell. A bacterium will move forward, called a “run,” when there is a CCW rotation, and reorient randomly, called a “tumble,” when there is a CW rotation.
Corkscrew Motility
Some spiral-shaped bacteria, known as the Spirochetes, utilize a corkscrew-motility due to their unusual morphology and flagellar conformation. These gram negative bacteria have specialized flagella that attach to one end of the cell, extend back through the periplasm and then attach to the other end of the cell. When these endoflagella rotate they put torsion on the entire cell, resulting in a flexing motion that is particularly effective for burrowing through viscous liquids.
Gliding Motility
Gliding motility is just like it sounds, a slower and more graceful movement than the other forms covered so far. Gliding motility is exhibited by certain filamentous or bacillus bacteria and does not require the use of flagella. It does require that the cells be in contact with a solid surface, although more than one mechanism has been identified. Some cells rely on slime propulsion, where secreted slime propels the cell forward, where other cells rely on surface layer proteins to pull the cell forward.
Chemotaxis
Now that we have covered the basics of the bacterial flagellar motor and mechanics of bacterial swimming, let us combine the two topics to talk about chemotaxis or any other type of taxes (just not my taxes). Chemotaxis refers to the movement of an organism towards or away from a chemical. You can also have phototaxis, where an organism is responding to light. In chemotaxis, a favorable substance (such as a nutrient) is referred to as an attractant, while a substance with an adverse effect on the cell (such as a toxin) is referred to as a repellant. In the absence of either an attractant or a repellant a cell will engage in a “random walk,” where it alternates between tumbles and runs, in the end getting nowhere in particular. In the presence of a gradient of some type, the movements of the cell will become biased, resulting over time in the movement of the bacterium towards an attractant and away from any repellants. How does this happen?
First, let us cover how a bacterium knows which direction to go. Bacteria rely on protein receptors embedded within their membrane, called chemoreceptors, which bind specific molecules. Binding typically results in methylation or phosphorylation of the chemoreceptor, which triggers an elaborate protein pathway that eventually impacts the rotation of the flagellar motor. The bacteria engage in temporal sensing, where they compare the concentration of a substance with the concentration obtained just a few seconds (or microseconds) earlier. In this way they gather information about the orientation of the concentration gradient of the substance. As a bacterium moves closer to the higher concentrations of an attractant, runs (dictated by CCW flagellar rotation) become longer, while tumbling (dictated by CW flagellar rotation) decreases. There will still be times that the bacterium will head off in the wrong direction away from an attractant since tumbling results in a random reorientation of the cell, but it won’t head in the wrong direction for very long. The resulting “biased random walk” allows the cell to quickly move up the gradient of an attractant (or move down the gradient of a repellant).
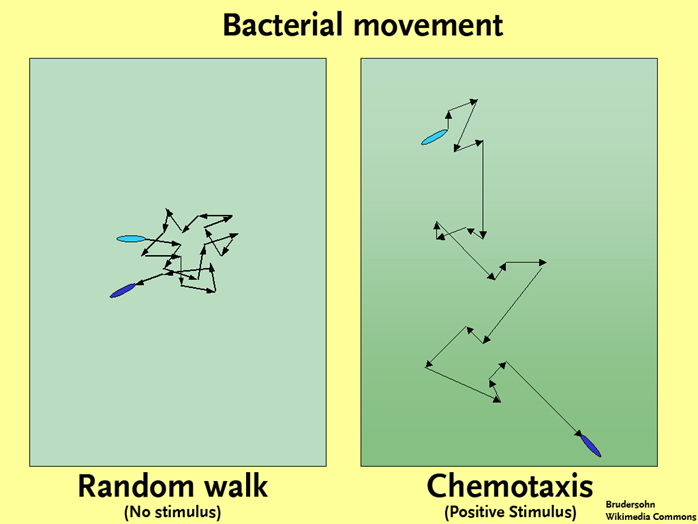
Bacterial Movement. By Brudersohn (Own work (Original text: selbst erstellt)) [CC BY-SA 2.0 de], via Wikimedia Commons
Key Words
capsule, slime layer, S-layer, fimbriae/fimbria, pilin, pili/pilus, conjugative pili, conjugation, type IV pili, twitching motility, flagella/flagellum, filament, flagellin, hook, motor, basal body, stator, Mot proteins, swimming, clockwise (CW), counterclockwise (CCW), run, tumble, Spirochetes, corkscrew-motility, endoflagella, gliding motility, chemotaxis, phototaxis, attractant, repellant, random walk, chemoreceptors, temporal sensing, biased random walk.
Essential Questions/Objectives
- What are the compositions and functions of capsules and slime layers? When are they produced? How do capsules or slime layers increase the survival chances of bacteria in different environments?
- What are fimbriae and pili; what are their compositions and functions?What is the size of bacterial flagella and how can they be arranged on a bacterial cell? How common are flagella in bacteria?
- What is the basic composition of a bacterial flagellum and how does this differ from flagella found in eukaryotes? How do bacterial flagella grow and how are proteins transported across the membrane? How do they cause movement? How is the movement different from Eukaryotic flagella?
- How are bacterial flagella attached to the body? How do the 2 inner rings work to cause movement and what powers the movement? What is the purpose of the 2 outer rings found in the basal body of gram-bacteria? What do gram + have instead?
- How do endoflagella differ from flagella and in what type of bacteria are they found? Where do they work better than flagella?
- What is chemotaxis? How does the direction of rotation of the flagella affect the way a bacteria moves? What do we know about the mechanism of chemotaxis in terms of membrane binding-proteins and chemotactic mediator? How long do stimuli last in chemotaxis and why is this important to the phenomenon?
Exploratory Questions (OPTIONAL)
- How could chemotaxis in microbes be used to address environmental pollution problems?


