22.1: Introduction
- Page ID
- 41050
In recent years, many subtle and previously disregarded mechanisms for fine genetic regulation have been discovered. Aside from direct regulation by proteins, these mechanisms include the involvement of non- protein coding regions of the genome, epigenomic factors such histone modifications, and diverse RNA switches. The spatial organization of chromatin inside the nucleus, chromatin modifier complexes and its functional consequences have also become an area of interest. In this chapter, we will delve into the study of 3D chromatin structures, starting with the state of art in this field, the most relevant terminology and current methods. Specially we will focus on the study of DNA regions located by peripheral regions of the nucleus (thus in close contact with the nuclear lamina) Finally, we will discuss the computational methods involved in studying nuclear genome organization.
What’s already known
DNA is locally compacted in nucleosomes, by wrapping around histone octamers. Each nucleosome comprises about 147 bps packed in 1.67 lef-handed superhelical turns. DNA is globally compacted as chromosomes (during cell division and mitosis). Chromosomes have been dyed with di↵erent colors, and it has been shown that some chromosomes have radial preferences within the cell nucleus, even when the cell is not actively undergoing division and the chromosomes are not condensed. That is, some chromosomes prefer to stay near the center of the nucleus while others tend towards the periphery. These are known as chromosome territories (CT). The territories of homologous chromosomes usually do not lie not near one another. It is also know that there is an overarching nuclear ’architecture’ that is observable, conserved even between different cell types.
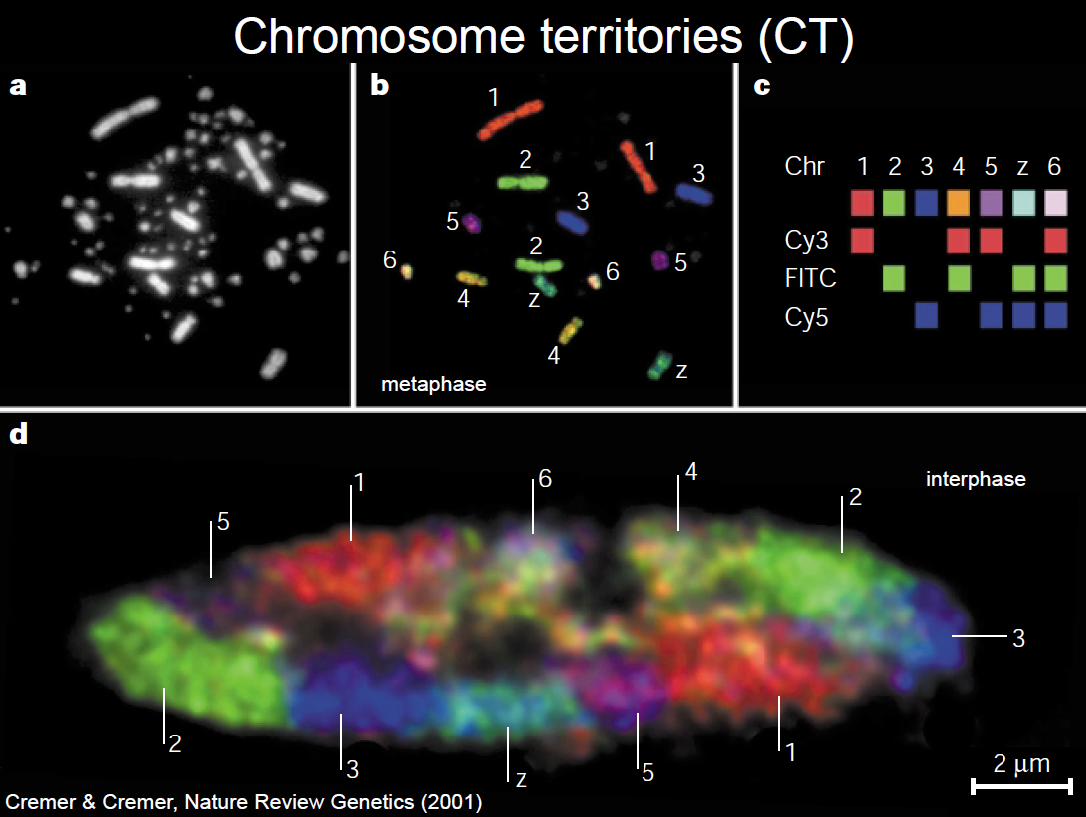
Courtesy of Macmillan Publishers Limited. Used with permission.
Source: Cremer, Thomas, and Christoph Cremer. "Chromosome Territories, Nuclear Architecture
and Gene Regulaiton in Mammalian Cells." Nature Reviews Genetics 2, no.4 (2001): 292-301.
What we don’t know
While local organization (nucleosome packaging) and global organization (chromosome condensation) of DNA are somehow understood, the intermediate structures of DNA are not yet well characterized - many speculated states have only been observed in vitro. The positioning of genomic regions in the nucleus on a sub-chromosomal level, for example the specific 3D conformation of a certain chromosomal region containing several genes, is also largely unknown.
While it is known that chromosomes retain some general architecture during the entire cell cycle, it is unknown how that location is maintained and how di↵erent chromosomes continue to interact over the course of the entire cell cycle.
Together, although we do understand certain parts of the function of chromosomes, we do not have a complete mechanistic understanding of this process.
Why do we study it?
In general, we are interested in understanding the functional characteristics of genomic regions and the molecular mechanisms encoded within, which might have implications in human diseases. Particularly, it has been shown that genes that are encoded in spatially neighboring regions are likely to be co-regulated. Also, the DNA packed inside the nucleus is the equivalent of wrapping 20 km of 20 μm thick thread in something the size of a tennis ball, which would reach from Kendall Square to Harvard and back over 6 and a half times! Isn't this amazing??


