B. MP2: An Overview of Metabolic Pathways - Anabolism
- Page ID
- 4616
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Anabolism: Anabolic reactions are those that lead to the synthesis of biomolecules. In contrast to the catabolic reactions just discussed (glycolysis, TCA cycle and electron transport/oxidative phosphorylation) which lead to the oxidative degradation of carbohydrates and fatty acids and energy release, anabolic reactions lead to the synthesis of more complex biomolecules including biopolymers (glycogen, proteins, nucleic acids) and complex lipids. Many biosynthetic reactions, including those for fatty acid synthesis, are reductive and hence require reducing agents. Reductive biosynthesis and complex polymer formation require energy input, usually in the form of ATP whose exergonic cleavage is coupled to endergonic biosynthesis.
Cells have evolved interesting mechanism so as not to have oxidative degradation reactions (which release energy) proceed at the same time and in the same cell as reductive biosynthesis (which requires energy input). Consider this scenario. You dive into a liver cell and find palmitic acid, a 16C fatty acid. From where did it come? Was it just synthesized by the liver cell or did it just enter the cell from a distant location such as adipocytes (fat cells). Should it be oxidized, which should happen if there is a demand for energy production by the cell, or should the liver cell export it, perhaps to adipocytes, which might happen if there is an excess of energy storage molecules? Cells have devised many ways to distinguish these opposing needs. One is by using a slightly different pool of redox reagents for anabolic and catabolic reactions. Oxidative degradation reactions typically use the redox pair NAD+/NADH (or FAD/FADH2) while reductive biosynthesis often uses phosphorylated variants of NAD+, NADP+/NADPH. In addition, cells often carry out competing reactions in different cellular compartments. Fatty acid oxidation of our example molecule (palmitic acid) occurs in the mitochondrial matrix, while reductive fatty acid synthesis occurs in the cytoplasm of the cell. Fatty acids entering the cell destined for oxidative degradation are transported into the mitochondria by the carnitine transport system. This transport system is inhibited under conditions when fatty acid synthesis is favored. We will discuss the regulation of metabolic pathways in a subsequent section. One of the main methods, as we will see, is to activate or inhibit key enzymes in the pathways under a given set of cellular conditions. The key enzyme in fatty acid synthesis, acetyl-CoA carboxylase, is inhibited when cellular conditions require fatty acid oxidation.
The following examples give short descriptions of anabolic pathways. Compare them to the catabolic pathways from the previous section.
-
Glucose synthesis, better known as Gluconeogenesis: In glycolysis, glucose (C6H12O6), a 6C molecule, is converted to two, 3C molecules (pyruvate) in an oxidative process that requires NAD+ and makes two net ATP molecules. In a few organs, most predominately in the liver, the reverse pathway can take place. The liver does this to provide glucose to the brain when the body is deficient in circulating glucose, for example, under fasting and starving conditions. (The liver under these conditions can get its energy from oxidation of fatty acids). The reactions in gluconeogenesis are the same reactions in glycolysis but run in reverse, with the exception of three glycolytic steps which are essentially irreversible. These three steps have bypass enzymes in the gluconeogenesis pathway. Although the synthesis of glucose is a reductive pathway, it uses NADH instead of NADPH as the redundant as the same enzyme used in glycolysis is simply run in reverse. Gluconeogenesis, which also occurs in the cortex of the kidney, is more than just a simple reversal of glycolysis, however. It can be thought of as the net synthesis of glucose from non-carbohydrate precursors. Pyruvate, as seen in the section on catabolism, can be formed from protein degradation to glucogenic amino acids which can be converted to pyruvate. It can also be formed from triacylglycerides from the 3C molecule glycerol formed and released from adipocytes after hydrolysis of three fatty acids from triacylglycerides. However, in humans, glucose can not be made in net fashion from fatty acids. Fatty acids can be converted to acetyl-CoA by fatty acid oxidation. The resulting acetyl-CoA can not form pyruvate since the enzyme that catalyzes the formation for acetyl-CoA from pyruvate, pyruvate dehydrogenase, is irreversible and there is no bypass reaction known. The acetyl-CoA can enter the TCA cycle but since the pathway is cyclic and proceeds in one direction, it can not form in net fashion oxaloacetate. Although oxaloacetate can be remove from the TCA cycle and be use to form phosphoenolpyuvate, a glycolytic intermediate, one acetyl-CoA condenses with one oxaloacetate to form citrate which leads back to one oxaloacetate. Hence fatty acids can not be converted to glucose and other sugars in a net fashion.
Figure: Gluconeogenesis
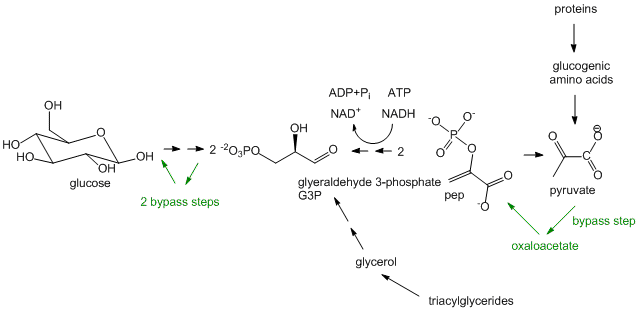
-
Pentose Phosphate Shunt: This two-part pathway doesn't appear to start as a reductive biosynthetic pathway as the first part is the oxidative conversion of a glycolytic intermediate, glucose-6-phosphate, to ribulose-5-phosphate. The next, nonoxidative branch leads to the formation of ribose-5-phosphate, a key biosynthetic intermediate in nucleic acid synthesis as well as erthyrose-4-phosphate used for biosynthesis of aromatic amino acids . The oxidative branch is important in reductive biosynthesis as it is a major source of the reductant NADPH used in biosynthetic reactions.
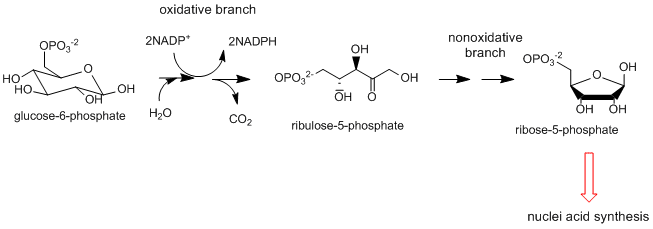
-
Fatty acid and isoprenoid/sterol biosynthesis: Acetyl-CoA is the source of carbon atoms for the synthesis of more complex lipids such as fatty acids, isoprenoids, and sterols. When energy needs in a cell are not high, citrate, the condensation product of oxaloacetate and acetyl-CoA in the TCA cycle, builds up in the mitochondrial matrix. It is then transported by the citrate transporter (an inner mitochondrial membrane protein) to the cytoplasm, where it is cleaved back to oxaloacetate and acetyl-CoA by the cytoplasmic enzyme citrate lyase. The oxaloacetate is returned to the mitochondria by conversion first to malate (reduction reaction using NADH), which can move back into the mitochondria through the malate transporter, or further conversion to pyruate, using the cytosolic malic enzyme, which uses NADP+ to oxidize malate to pyruvate which then enters the mitochondria. The acetyl-CoA formed in the cytoplasm can then be used in reductive biosynthesis using NADPH as the reductant to form fatty acids, isoprenoids, and sterols. The NADPH for the reduction comes from the oxidative branch of the pentose phosphate pathway and from the reaction catalyzed by malic enzyme. The liver cells can still run the glycolytic pathway as the NADH/NAD+ ratio is low in the cytoplasm while NADPH/NADP+ ratio is high.
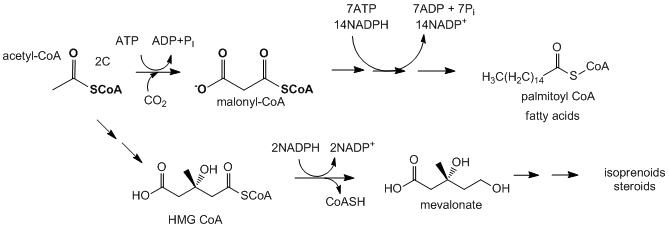
Now its time to see how the various pathways fit together to form an integrated set of pathways.


