1.6: Lipids and Signaling
- Page ID
- 4530
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Learning Objectives
- Describe how lipid bilayers participate in signal transduction
F1. Introduction to Lipid Signaling
Swedish Translation √ by Valeria Aleksandrova
Lipids are not just used as a passive component of membranes, or as a source of stored energy. They are involved in the process of signal transduction at the cell membrane, a process by which the interior components of the cell respond to a signal external to the cell, allowing the cell to respond to their local environment. Usually a chemical signal on the outside of the cell is the "primary messenger" that causes the cell to respond. Usually the chemical transmitter of information does not get into the cell. Rather it binds to surface receptors on the cell membrane surface. Somehow, the cells senses that a ligand is bound to the outside. Enzymes, usually in the membrane or at the intracellular surface of the lipid bilayer are activated. Many of these enzymes cleave lipids in the membrane. The cleaved fragments of the lipid molecules serve as intracellular signals or "secondary messengers" , which can bind to intracellular enzymes to activate intracellular processes. The following diagram shows some of the lipid mediators which are generated by the process and signal the cell to respond.

Figure: Lipids - More than Grease - Mediators in Signal Transduction
Recently, fatty acid amides have been shown to be potent mediators of neurological processes. In one interesting experiment, sheep were sleep deprived. Reasoning that the brain might release a biochemical signal into cerebrospinal fluid to induce sleep, scientists at Scripps removed some of this fluid and isolated a substance that was not found in rested sheep. On analysis, the structure was shown to be an amide of oleic acid.. Oleylethanolamide has been shown to bind to the peroxisome-proliferator-activated receptor-a (PPAR-a) which resides in the nucleus. This ligand, by affecting gene transcription, appears to regulate body weight and the feeling of fullness after eating (satiety) as it ieads to reduced eating.
In an analogous fashion, people have sought the natural neurotransmitter which binds to the same receptor in the brain as THC, the active ingredient of marijuana. This was found several years ago and was shown to be the amide of arachidonic acid, called anandamide. See the figure below for structures.
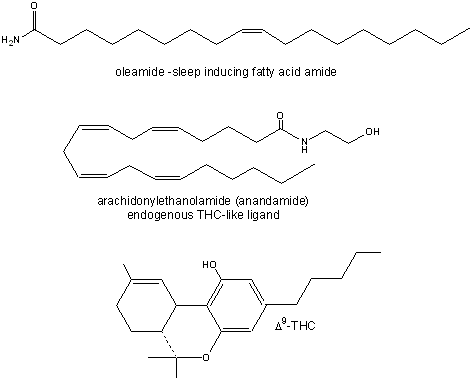
Figure: Fatty Acid Amides: Neurochemical Mediators
This fatty acid amide is an examples of a class of lipid derivatives called N-acylethanolamines (NAEs). These molecules, with acyl groups that vary in number of carbons and double bonds, are found widely in organisms in nature. Naturally occurring anandamide leads to increased food intake after a short period of reduced food intake. One of the known physiological effects of THC is increased food consumption (the munchies).
In a recent study, Lucanic et al (2011) have shown that decreases in NAEs extend the the life span in the small roundworm C. elegans, which has become a model organism to study genes in eukaryotes. Caloric restriction has been shown to increase life span in a variety of organisms. In invertebrates, anandamide seems to inhibit food intake, even in organisms that lack a receptor similar to which cannabinoids binds. This might seem paradoxical in that anandamide (and THC) in humans seems to induce eating. However, under long periods of caloric restriction (low level starvation) in rats, anandamide levels are suppressed, leading to a low energy consuming state.
In generally it appears that reductions of NAEs occur during periods of caloric restriction. Mutant worms which have reduced levels of NAEs through targeted enzyme disruptions that affected either NAE synthesis or degradation have longer life spans. If normal (wild-type) worms were placed under caloric restriction but given EPA-ethanolamine (the most abundant NAE in these worms), the did not have an extended lifespan.
References
- Lucanic, N. et al. N-acylethanolamine signaling mediates the effect of diet on lifespan in Caenorhabditis elegans. Nature, 473, 226-229 (2011).


