D1. Introduction to Transcription
- Page ID
- 4976
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
Imagine you have been given a string approximately 2 meters long, which represents an unwound, deproteinated, human chromosome. In actuality, such naked DNA does not exist in the cell nucleus. Rather, it is wound around a series of positively charged histone proteins. In the electron microscope it resembles beads on a string. This structure is wound into a cylindrical "solenoid" structure which is further packaged to fit into the nucleus along with the rest of the chromosomes. There just happens to be a small dot in the dead center of the string you have been given. It represents the gene for a particular protein called metallothionein. This gene is expressed and the protein metallothionein is made when cells are exposed to heavy metals like Cd. How might the cell not express the gene, or express it to a small, constitutive level, in the absence of Cd exposure? How might the gene might be activated - through transcription of the gene and translation of the resulting mRNA to form the metallothionein protein upon exposure to Cd. (Hint: Cd doe not DIRECTLY bind to DNA.)?
One of the central questions of modern biology is what controls gene expression. As we have previously described, genes must be "turned on" at the right time, in the right cell. To a first approximation, all the cells in an organism contains the same DNA (with the exception of germ cells and immune cells). Cell type is determined by what genes are expressed at a given time. Likewise, cell can change (differentiate) into different types of cells by altering the expression of genes. The Central Dogma of Biology describes how genes are first transcribed to messenger RNA (mRNA), and then the mRNA is translated into a corresponding protein sequence. The links below should be reviewed by those who have little background on the Central Dogma of Biology and of the nature of a gene.
- overview of Central Dogma of Biology for the non-biochemistry major
- A view of genes and their products: Simplicity to Complexity
![]() Updated Simple DNA Tutorial Jmol14 (Java) | JSMol (HTML5)
Updated Simple DNA Tutorial Jmol14 (Java) | JSMol (HTML5)
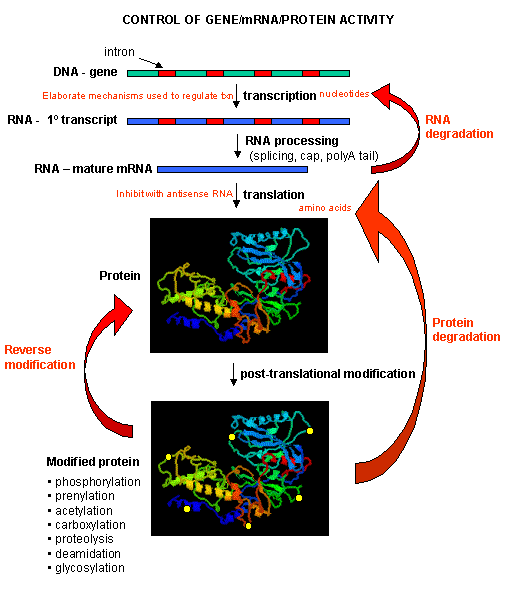
Once made, proteins can then be post-translationally modified, localized to certain sites within the cells, and ultimately degraded. If functional proteins are considered the end-product of gene expression, the control of gene expression could theoretically occur at any of these steps in the process.
Figure: PROCESSES THAT AFFECT THE STEADY STATE CONCENTRATION OF A PROTEIN
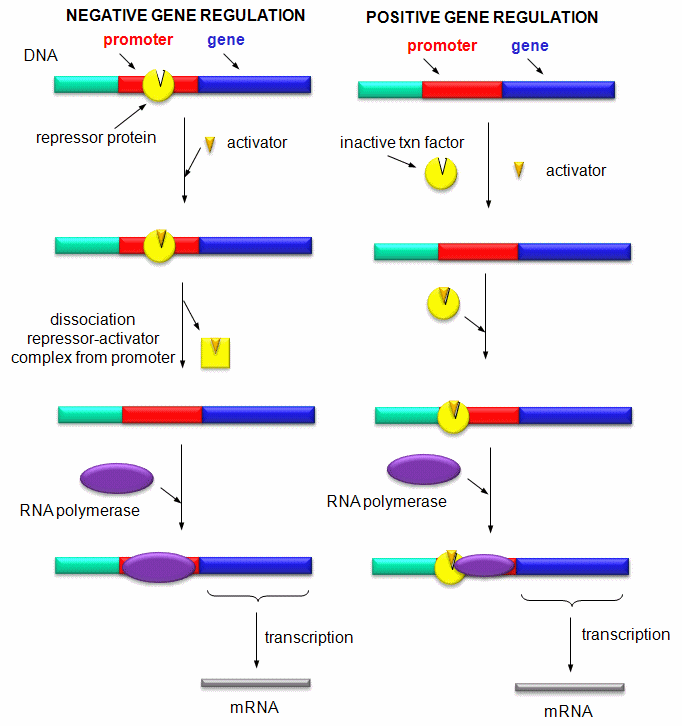
Mostly, however, gene expression appears to be controlled at the level of transcription. This makes great biological sense, since it would be less energetically wasteful to induce or inhibit the ultimate expression of a functional protein at a step early in the process. How can gene expression be regulated at the transcriptional level? Many examples have been documented. The main control is typically exerted at the level of RNA polymerase binding just upstream (5') of a site for transcriptional initiation. Other factors, called transcription factors (which are usually proteins), bind to the same region and promote the binding of RNA polymerase at its binding site, called the promoter. Proteins can also bind to sites on DNA (operator in prokaryotes) and inhibit the assembly of the transcription complex and hence transcription. Regulation of gene transcription then becomes a matter of binding the appropriate transcription factors and RNA polymerase to the appropriate region at the start site for gene transcription. Regulation of gene expression by proteins can be either positive or negative. Regulation in prokaryotes is usually negative while it is positive in eukaryotes.
Figure: Positive and Negative Regulation of Gene Transcription