10.3: Genome editing
- Page ID
- 148622
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Thus far, we’ve only discussed transgenes that are either randomly inserted into the genome or don’t interact with the genome at all. However, if you want a durable cure to a genetic disease, then you want a genetic change to be heritable – and that means you need to make a change to the genome. And what you really want is to be able to say “I want THIS change made right HERE.” That’s the idea behind genome editing.
Engineering double-stranded breaks
Zinc-finger nucleases
How do you point at a particular piece of DNA and say “I want a change here please?” There are three tools that are commonly used. The first are called zinc-finger nucleases – nucleases that contain a series of zinc finger domains that each recognizes a three nucleotide sequence fused to a nuclease domain. A number of zinc finger domains have been described, and you can swap them in and out to recognize between 9 and 18 bases, though not every possible set of bases is recognizable and specificity is problematically low. When the ZFN finds the site in the genome, it cuts the DNA there.

TAL-effector nucleases (TALENs)
The second tool is called a TAL-effector nuclease, or TALEN. These are also “programmable” nucleases, insofar as you can build a nuclease that recognizes a specific sequence by combining multiple protein domains that each recognize a few bases. And again, these recognition domains are fused to a nuclease domain that cuts the genome at that location.

CRISPR
The most recent is CRISPR. CRISPR is an umbrella term for a set of programmable nucleases – the most famous is called Cas9. Cas9 binds to a short RNA called a “guide RNA” and uses this RNA to find the site in the genome to cut. CRISPR is exciting because it doesn’t require engineering a new nuclease each time you want to direct it to a different spot in the genome. Instead, just synthesize a guide RNA to “reprogram” the nuclease.

The results of double-stranded breaks
Non-homologous end-joining
In each case, a ZFN or a TALEN or a CRISPR nuclease creates a double-stranded break in the genome. Why is this helpful? Well, sometimes a therapy might want to disable a gene instead of add one. For example, haemophilia B is a dominant disorder – if you cause a DSB in the middle of the gene, then the cell joins the ends back together with a process called non-homologous end joining, or NHEJ. The cell basically “sticks” the broken ends back together, and that usually the repair loses a base or two. This introduces a frameshift mutation into the gene that disables it.
Homology-directed repair (HDR)
A double-stranded break in a chromosome is a big problem for a cell – when the cell divides, it’s not going to be able to completely copy that chromosome into daughter cells. As a result, there are two common ways a cell tries to repair it. One we’ve already seen, which is non-homologous end joining – the cell sticks the two broken ends back together. Often the join isn’t perfect, and if it’s in the middle of a gene a mutation gets introduced which often disables the gene.
However, if there’s a “non-broken” copy of the chromosome around – say, the cell is after S phase and there’s another chromatid right next door – then the cell can use the working copy to repair the damaged one. This process is called non-allelic homologous recombination, and importantly it depends on homologous regions before and after the break. Gene therapies – and other genetic engineering – can use NAHR to precisely modify the genome by stimulating a double-stranded break with a nuclease, then providing a “working copy” for the cell to repair the DSB with. If the break is precisely positioned at a disease-causing mutation, then the “working copy” can repair that mutation.

Base editing
Making double-stranded breaks to stimulate non-allelic homologous repair is still a risky business, though. Despite being “programmable”, these nucleases often cut places they’re not supposed to and can cause other DNA damage. A new generation of ultra-precise nucleases does a better job, but – what if we could repair mutations without a double-stranded break? What if we replace the DNA-cutting domain with some other functional domain? Then, we would have a protein that can bring that functional domain to a specified sequence of DNA in the genome.
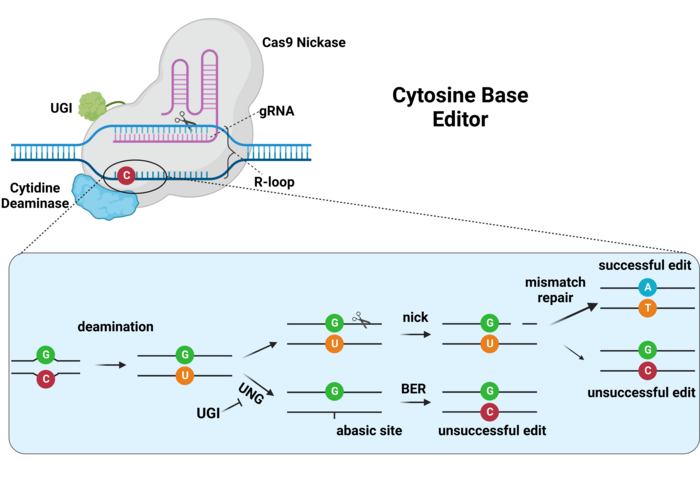
This is the idea behind base editing. 58% of the human genetic variants associated with disease are point mutations, and a base-editor can change a C-G pair to a T-A pair or vice-versa. Here’s how this works for a C to T edit. A Cas9 CRISPR nuclease is changed so that instead of cutting DNA, it nicks it instead. We’ll call this a “nicking Cas9”, or “nCas9”. To the nCas9, we’ll fuse a cytidine deaminase and program it with a guide RNA that brings the nCas9 to the site we want to repair. The deaminase turns the cytosine into a uracil and the nCas9 nicks the other strand a few bases downstream. This nick causes the cell to repair the nick, and that process chews back a few bases then synthesizes the complementary strand. In this case, however, the complementary base to a uracil is a thymine! Then, DNA replication or further repair processes recognize the uracil and replace it with an adenine. Neat, huh? Changing a C-G to an A-T works similarly. Base editing is not used in any approved therapies yet, but they’re sufficiently precise that a number of therapies are in phase I and phase II clinical trials.