Electron Transport Chains*#
- Page ID
- 8166
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Electron Transport Chains
An electron transport chain, or ETC, is composed of a group of protein complexes in and around a membrane that help energetically couple a series of exergonic/spontaneous redox reactions to the endergonic pumping of protons across the membrane to generate a an electrochemical gradient. This electrochemical gradient creates a free energy potential that is termed a proton motive force whose energetically "downhill" exergonic flow can later be coupled to a variety of cellular processes.
ETC Overview
Step 1. Electrons enter the ETC from an electron donor, such as NADH or FADH2 which are generated during a variety of catabolic reactions, like and including those associated glucose oxidation . Depending on the complexity (number and types of electron carriers) of the ETC being used by an organism, electrons can enter at a variety of places in the electron transport chain - this depends upon the respective reduction potentials of the proposed electron donors and acceptors.
Step 2. After the first redox reaction, the initial electron donor will become oxidized and the electron acceptor will become reduced. The difference in redox potential between the electron acceptor an donor is related to ΔG by the relationship ΔG = -nFΔE, where n = the number of electrons transferred and F = Faraday's constant. The larger a positive ΔE the more exergonic a reaction.
Step 3. If sufficient energy transferred during an exergonic redox step the electron carrier may couple this negative change in free energy to the endergonic process of transporting a proton from one side of the membrane to the other.
Step 4. After multiple redox transfers, the electron is delivered to a molecule known as the terminal electron acceptor. In the case of humans, the terminal electron acceptor is oxygen. However, there are many, many, many, other possible electron acceptors, see below.
Note
Electrons entering the ETC do not have to come from NADH or FADH2. Many other compounds can serve as electron donors, the only requirements are that there exists an enzyme that can oxidize the electron donor and then reduce another compound and that the E0' be positive (e.g. ΔG<0). Even a small amounts of free energy transfers can add up. For example there are bacteria that use H2 as an electron donor. This is not too difficult to believe because the half reaction 2H+ + 2 e-/H2 has a reduction potential (E0') of -0.42 V. If these electrons are eventually delivered to oxygen then the ΔE0' of the reaction is 1.24 V which corresponds to a large negative ΔG (-ΔG). Alternatively, there are some bacteria that can oxidize iron, Fe2+ at pH 7 to Fe3+ with a reduction potential (E0') of + 0.2 V. These bacteria use oxygen as their terminal electron acceptor and in this case, the ΔE0' of the reaction is approximately 0.62 V. This still produces a -ΔG. The bottom line is that depending on the electron donor and acceptor that the organism uses, a little or a lot of energy can be transferred and used by the cell per electrons donated to the electron transport chain.
What are the complexes of the ETC?
ETCs are made up of a series (at least one) of membrane associated redox proteins or (some are integral) protein complexes (complex = more than one protein arranged in a quaternary structure) that move electrons from a donor source, such as NADH, to a final terminal electron acceptor, such as oxygen. This particular donor/terminal acceptor pair is the primary one used in human mitochondria. Each electron transfer in the ETC requires a reduced substrate as an electron donor and an oxidized substrate as the electron acceptor. In most cases the electron acceptor is a member of the enzyme complex. Once the complex is reduced, the complex can serve as an electron donor for the next reaction.
How do ETC complexes transfer electrons?
As previously mentioned the ETC is composed of a series of protein complexes that undergo a series of linked redox reactions. These complexes are in fact multiprotein enzyme complexes referred to as oxidoreductases or simply reductases. The one exception to this naming convention is the terminal complex in aerobic respiration that uses molecular oxygen as the terminal electron acceptor. That enzyme complex is referred to as an oxidase. Redox reactions in these complexes are typically carried out by a non-protein moiety called a prosthetic group. This is true for all of the electron carriers with the exception of quinones, which are a class of lipids that can directly be reduced or oxidized by the oxidoreductases. In this case, both the Quinonered and the Quinoneox is soluble within the membrane and can move from complex to complex. The prosthetic groups are directly involved in the redox reactions being catalyzed by their associated oxidoreductases. In general these prosthetic groups can be divided into two general types: those that carry both electrons and protons and those that only carry electrons.
The Electron and Proton carriers
- Flavoproteins (Fp), these proteins contain an organic prosthetic group called a flavin, which is the actual moiety that undergoes the oxidation/reduction reaction. FADH2 is an example of a Fp.
- Quinones, are a family of lipids which means they are soluble within the membrane.
- It should also be noted that NADH and NADPH are considered electron (2e-) and proton (2 H+) carriers.
Electon carriers
- Cytochromes are proteins that contain a heme prosthetic group. The Heme is capable of carrying a single electron.
- Iron-Sulfur proteins contain a non-heme iron-sulfur clusters that can carry an electron. The prosthetic group is often abbreviated as Fe-S
Aerobic versus Anaerobic respiration
In the world we live in, most of the organisms we interact with (at least those we see) breath air, which is approximately 20% oxygen. Oxygen is our terminal electron acceptor. We call this process respiration, specifically aerobic respiration. We breath in oxygen, our cells take it up and transport it into the mitochondria where it is used as the final acceptor of electrons from our electron transport chains. That is aerobic respiration: the process of using oxygen as a terminal electron acceptor in an electron transport chain.
While we may use oxygen as the terminal electron acceptor for our respiratory chains, the more general process of respiration evolved at time when oxygen was not a major component of the atmosphere. Respiration or oxidative phosphorylation does not require oxygen at all; it simply requires a compound with a high reduction potential to act as a terminal electron acceptor; accept electrons from one of the complexes within the ETC. Many organisms can use a variety of compounds including nitrate (NO3-), nitrite (NO2-), even iron (Fe+++) as terminal electron acceptors. When oxygen is NOT the terminal electron acceptor, the process is referred to as anaerobic respiration. The ability of organisms to vary their terminal electron acceptor provides metabolic flexibility and can ensure better survival if any given terminal acceptor is in limited supply. Think about this, in the absence of oxygen we die; but an organism that can use a different terminal electron acceptor can survive.
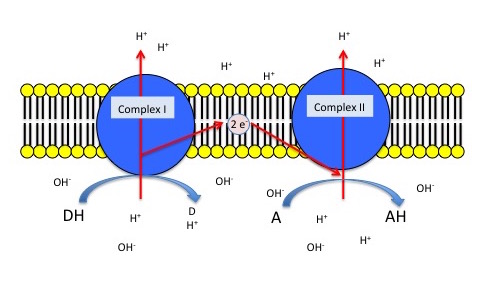
A generic example of a simple, 2 complex ETC
The figure below depicts a generic electron transport chain, composed of two integral membrane complexes; Complex Iox and Complex IIox. A reduced electron donor, designated DH (such as NADH or FADH2) reduces Complex Iox giving rise to the oxidized form D (such as NAD+ or FAD+). Simultaneously, a prosthetic group within complex I is now reduced (accepts the electrons). In this example the redox reaction is exergonic and the free energy difference is coupled by the enzymes in Complex I to the endergonic translocation of a proton from one side of the membrane to the other. The net result is that one surface of the membrane becomes more negatively charged, due to an excess of hydroxyl ions (OH-) and the other side becomes positively charged due to an increase in protons on the other side. Complex Ired can now reduce the prosthetic group in Complex IIred while simultaneously oxidizing Complex Ired. Electrons pass from Complex I to Complex II via thermodynamically spontaneous redox reactions, regenerating Complex Iox which can repeat the previous process. Complex IIred reduces A, the terminal electron acceptor to regenerate Complex IIox and create the reduced form of the terminal electron acceptor. In this case, Complex II can also translocate a proton during the process. If A is molecular oxygen, water (AH) will be produced. When A is oxygen the reaction scheme would be considered a model of an aerobic ETC. However, if A is nitrate, NO3- then Nitrite, NO2- is produced (AH) and this would be an example of an anaerobic ETC.
Generic 2 complex electron transport chain. In the figure, DH is the electron donor (donor reduced) and D is the donor oxidized. A is the oxidized terminal electron acceptor and AH is the final product, the reduced form of the acceptor. As DH is oxidized to D, protons are translocated across the membrane, leaving an excess of hydroxyl ions (negatively charged) on one side of the membrane and protons (positively charged) on the other side of the membrane. The same reaction occurs in Complex II as the terminal electron acceptor is reduced to AH.
Exercise 1
Thought question
Based on the figure above and using an electron tower, what is the difference in the electrical potential if (A) DH is NADH and A is O2 and (B) DH is NADH and A is NO3-. Which pairs (A or B) provide the most amount of usable energy?
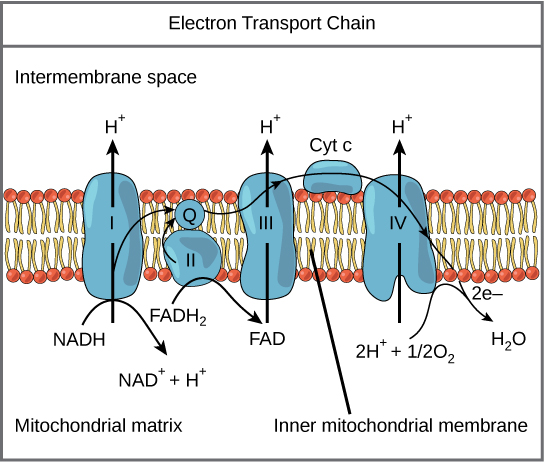
Detailed look at aerobic respiration
The eukaryotic mitochondria has evolved a very efficient ETC. There are four complexes composed of proteins, labeled I through IV depicted in the figure below. The aggregation of these four complexes, together with associated mobile, accessory electron carriers, is called also called an electron transport chain. This type of electron transport chain is present in multiple copies in the inner mitochondrial membrane of eukaryotes.
The electron transport chain is a series of electron transporters embedded in the inner mitochondrial membrane that shuttles electrons from NADH and FADH2 to molecular oxygen. In the process, protons are pumped from the mitochondrial matrix to the intermembrane space, and oxygen is reduced to form water.
Complex I
To start, two electrons are carried to the first complex aboard NADH. This complex, labeled I, is composed of flavin mononucleotide (FMN) and an iron-sulfur (Fe-S)-containing protein. FMN, which is derived from vitamin B2, also called riboflavin, is one of several prosthetic groups or co-factors in the electron transport chain. A prosthetic group is a non-protein molecule required for the activity of a protein. Prosthetic groups are organic or inorganic, non-peptide molecules bound to a protein that facilitate its function; prosthetic groups include co-enzymes, which are the prosthetic groups of enzymes. The enzyme in complex I is NADH dehydrogenase and is a very large protein, containing 45 amino acid chains. Complex I can pump four hydrogen ions across the membrane from the matrix into the intermembrane space, and it is in this way that the hydrogen ion gradient is established and maintained between the two compartments separated by the inner mitochondrial membrane.
Q and Complex II
Complex II directly receives FADH2, which does not pass through complex I. The compound connecting the first and second complexes to the third is ubiquinone (Q). The Q molecule is lipid soluble and freely moves through the hydrophobic core of the membrane. Once it is reduced, (QH2), ubiquinone delivers its electrons to the next complex in the electron transport chain. Q receives the electrons derived from NADH from complex I and the electrons derived from FADH2 from complex II, including succinate dehydrogenase. This enzyme and FADH2 form a small complex that delivers electrons directly to the electron transport chain, bypassing the first complex. Since these electrons bypass and thus do not energize the proton pump in the first complex, fewer ATP molecules are made from the FADH2 electrons. As we will see in the following section, the number of ATP molecules ultimately obtained is directly proportional to the number of protons pumped across the inner mitochondrial membrane.
Complex III
The third complex is composed of cytochrome b, another Fe-S protein, Rieske center (2Fe-2S center), and cytochrome c proteins; this complex is also called cytochrome oxidoreductase. Cytochrome proteins have a prosthetic group of heme. The heme molecule is similar to the heme in hemoglobin, but it carries electrons, not oxygen. As a result, the iron ion at its core is reduced and oxidized as it passes the electrons, fluctuating between different oxidation states: Fe++ (reduced) and Fe+++ (oxidized). The heme molecules in the cytochromes have slightly different characteristics due to the effects of the different proteins binding them, giving slightly different characteristics to each complex. Complex III pumps protons through the membrane and passes its electrons to cytochrome c for transport to the fourth complex of proteins and enzymes (cytochrome c is the acceptor of electrons from Q; however, whereas Q carries pairs of electrons, cytochrome c can accept only one at a time).
Complex IV
The fourth complex is composed of cytochrome proteins c, a, and a3. This complex contains two heme groups (one in each of the two cytochromes, a, and a3) and three copper ions (a pair of CuA and one CuB in cytochrome a3). The cytochromes hold an oxygen molecule very tightly between the iron and copper ions until the oxygen is completely reduced. The reduced oxygen then picks up two hydrogen ions from the surrounding medium to make water (H2O). The removal of the hydrogen ions from the system contributes to the ion gradient used in the process of chemiosmosis.
Helpful Link
YouTube Electron Transport Chain
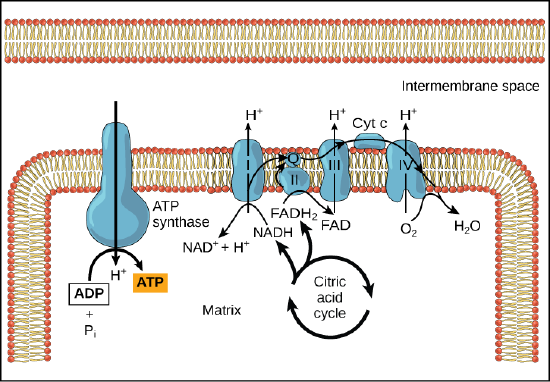
Chemiosmosis
In chemiosmosis, the free energy from the series of redox reactions just described is used to pump hydrogen ions (protons) across the membrane. The uneven distribution of H+ ions across the membrane establishes both concentration and electrical gradients (thus, an electrochemical gradient), owing to the hydrogen ions’ positive charge and their aggregation on one side of the membrane.
If the membrane were open to diffusion by the hydrogen ions, the ions would tend to diffuse back across into the matrix, driven by their electrochemical gradient. Many ions cannot diffuse through the nonpolar regions of phospholipid membranes without the aid of ion channels. Similarly, hydrogen ions in the matrix space can only pass through the inner mitochondrial membrane through an integral membrane protein called ATP synthase (depicted below). This complex protein acts as a tiny generator, turned by transfer of energy mediated by protons moving down their electrochemical gradient. The movement of this molecular machine (enzyme) serves to lower the activation energy of reaction and couples the exergonic transfer of energy associated with the movement of protons down their electrochemical gradient to the endergonic addition of a phosphate to ADP, forming ATP.
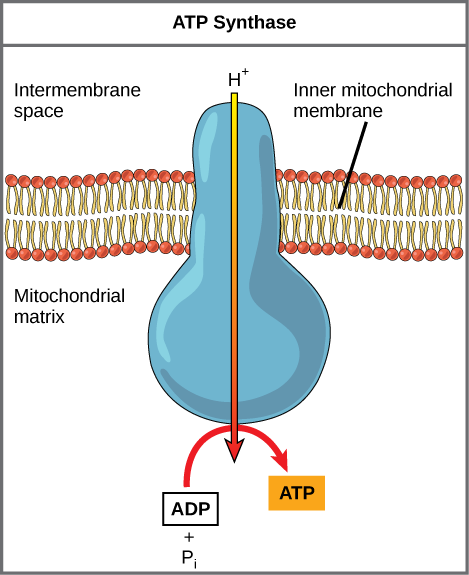
ATP synthase is a complex, molecular machine that uses a proton (H+) gradient to form ATP from ADP and inorganic phosphate (Pi). (Credit: modification of work by Klaus Hoffmeier)
Suggested Discussion
Dinitrophenol (DNP) is a small chemical that serves to uncouple the flow of protons across the inner mitochondrial membrane to the ATP synthase and thus the synthesis of ATP. DNP makes the membrane leaky to protons. It was used until 1938 as a weight-loss drug. What effect would you expect DNP to have on the difference in pH across both sides of the inner mitochondrial membrane? Why do you think this might be an effective weight-loss drug? Why might it be dangerous?
In healthy cells, chemiosmosis (depicted below) is used to generate 90 percent of the ATP made during aerobic glucose catabolism; it is also the method used in the light reactions of photosynthesis to harness the energy of sunlight in the process of photophosphorylation. Recall that the production of ATP using the process of chemiosmosis in mitochondria is called oxidative phosphorylation. The overall result of these reactions is the production of ATP from the energy of the electrons removed from hydrogen atoms. These atoms were originally part of a glucose molecule. At the end of the pathway, the electrons are used to reduce an oxygen molecule to water.
Links
How ATP is made from ATP synthase
In oxidative phosphorylation, the pH gradient formed by the electron transport chain is used by ATP synthase to form ATP in a Gram- bacteria.
Suggested Discussion
Cyanide inhibits cytochrome c oxidase, a component of the electron transport chain. If cyanide poisoning occurs, would you expect the pH of the intermembrane space to increase or decrease? What effect would cyanide have on ATP synthesis?