Transcription—from DNA to RNA*#
- Page ID
- 9428
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Section summary
Bacteria, archaea, and eukaryotes must all transcribe genes from their genomes. While the cellular location may be different (eukaryotes perform transcription in the nucleus; bacteria and archaea perform transcription in the cytoplasm), the mechanisms by which organisms from each of these clades carry out this process are fundamentally the same and can be characterized by three stages: initiation, elongation, and termination.
Transcription: from DNA to RNA
A short overview of transcription
Transcription is the process of creating an RNA copy of a segment of DNA. Since this is a process, we want to apply the Energy Story rubric to develop a functional understanding of transcription. What does the system of molecules look like before the start of the transcription? What does it look like at the end? What transformations of matter and transfers of energy happen during the transcription and what, if anything, catalyzes the process? We also want to think about the process from a Design Challenge standpoint. If the biological task is to create a copy of DNA in the chemical language of RNA, what challenges can we reasonably hypothesize or anticipate, given our knowledge about other nucleotide polymer processes, must be overcome? Is there evidence that Nature solved these problems in different ways? What seem to be the criteria for success of transcription? You get the idea.
Listing some of the basic requirements for transcription
Let us first consider the tasks at hand by using some of our foundational knowledge and imagining what might need to happen during transcription if the goal is to make an RNA copy of a piece of one strand of a double-stranded DNA molecule. We'll see that using some basic logic allows us to infer many of the important questions and things that we need to know in order to properly describe the process.
Let's imagine that we want to design a nanomachine/nanobot that would conduct transcription. We can use some Design Challenge thinking to identify problems and subproblems that need to be solved by our little robot.
• Where should the machine start? Along the millions to billions of base pairs, where should the machine be directed?
• Where should the machine stop?
• If we have start and stop sites, we will need ways of encoding that information so that our machine(s) can read this information—how will that be accomplished?
• How many RNA copies of the DNA will we need to make?
• How fast do the RNA copies need to be made?
• How accurately do the copies need to be made?
• How much energy will the process take and where is the energy going to come from?
These are, of course, only some of the core questions. One can dig deeper if they wish. However, these are already good enough for us to start getting a good feel for this process. Notice, too, that many of these questions are remarkably similar to those we inferred might be necessary to understand about DNA replication.
The building blocks of transcription
The building blocks of RNA
Recall from our discussion on the structure of nucleotides that the building blocks of RNA are very similar to those in DNA. In RNA, the building blocks consists of nucleotide triphosphates that are composed of a ribose sugar, a nitrogenous base, and three phosphate groups. The key differences between the building blocks of DNA and those of RNA are that RNA molecules are composed of nucleotides with ribose sugars (as opposed to deoxyribose sugars) and utilize uridine, a uracil containing nucleotide (as opposed to thymidine in DNA). Note below that uracil and thymine are structurally very similar—the uracil is just lacking a methyl (CH3) functional group compared to thymine.
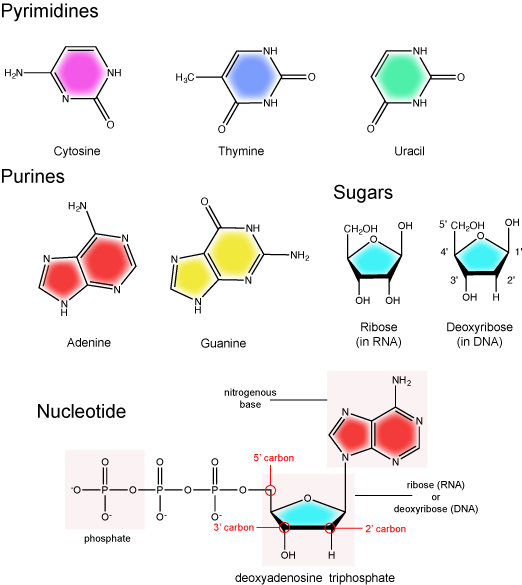
Figure 1. The basic chemical components of nucleotides.
Attribution: Marc T. Facciotti (original work)
Transcription initiation
Promoters
Proteins responsible for creating an RNA copy of a specific piece of DNA (transcription) must first be able to recognize the beginning of the element to be copied. A promoter is a DNA sequence onto which various proteins, collectively known as the transcription machinery, bind and initiates transcription. In most cases, promoters exist upstream (5' to the coding region) of the genes they regulate. The specific sequence of a promoter is very important because it determines whether the corresponding coding portion of the gene is transcribed all the time, some of the time, or infrequently. Although promoters vary among species, a few elements of similar sequence are sometimes conserved. At the -10 and -35 regions upstream of the initiation site, there are two promoter consensus sequences, or regions that are similar across many promoters and across various species. Some promoters will have a sequence very similar to the consensus sequence (the sequence containing the most common sequence elements), and others will look very different. These sequence variations affect the strength to which the transcriptional machinery can bind to the promoter to initiate transcription. This helps to control the number of transcripts that are made and how often they get made.
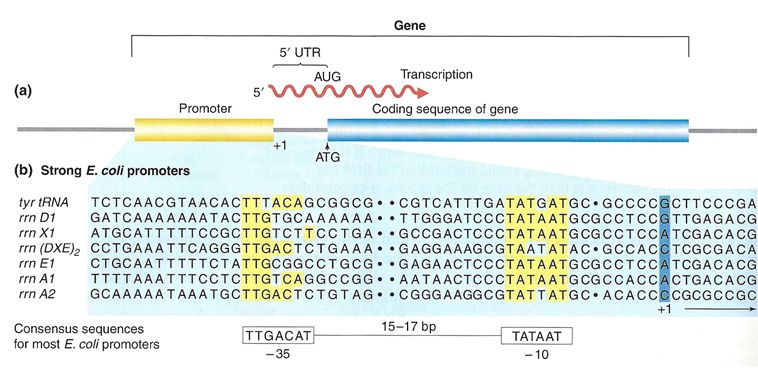
Figure 2. (a) A general diagram of a gene. The gene includes the promoter sequence, an untranslated region (UTR), and the coding sequence. (b) A list of several strong E. coli promoter sequences. The -35 box and -10 box are highly conserved sequences throughout the strong promoter list. Weaker promoters will have more base pair differences when compared to these sequences.
Source: http://www.discoveryandinnovation.co...lecture12.html
Note: possible discussion
What types of interactions are changed between the transcription machinery and the DNA when the nucleotide sequence of the promoter changes? Why would some sequences create a "strong" promoter and why do others create a "weak" promoter?
Bacterial vs. eukaryotic promoters
In bacterial cells, the -10 consensus sequence, called the -10 region, is AT rich, often TATAAT. The -35 sequence, TTGACA, is recognized and bound by the protein σ. Once this protein-DNA interaction is made, the subunits of the core RNA polymerase bind to the site. Due to the relatively lower stability of AT associations, the AT-rich -10 region facilitates unwinding of the DNA template, and several phosphodiester bonds are made.
Eukaryotic promoters are much larger and more complex than prokaryotic promoters, but both have an AT-rich region—in eukaryotes, it is typically called a TATA box. For example, in the mouse thymidine kinase gene, the TATA box is located at approximately -30. For this gene, the exact TATA box sequence is TATAAAA, as read in the 5' to 3' direction on the nontemplate strand. This sequence is not identical to the E. coli -10 region, but both share the quality of being AT-rich element.
Instead of a single bacterial polymerase, the genomes of most eukaryotes encode three different RNA polymerases, each made up of ten protein subunits or more. Each eukaryotic polymerase also requires a distinct set of proteins known as transcription factors to recruit it to a promoter. In addition, an army of other transcription factors, proteins known as enhancers, and silencers help to regulate the synthesis of RNA from each promoter. Enhancers and silencers affect the efficiency of transcription but are not necessary for the initiation of transcription or its procession. Basal transcription factors are crucial in the formation of a preinitiation complex on the DNA template that subsequently recruits RNA polymerase for transcription initiation.
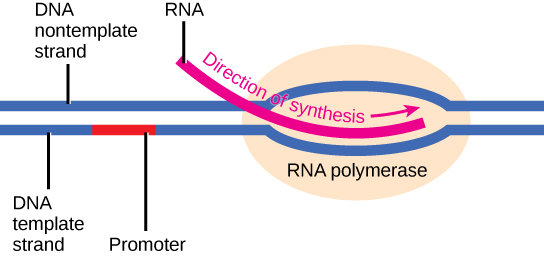
Initiation of transcription begins with the binding of RNA polymerase to the promoter. Transcription requires the DNA double helix to partially unwind such that one strand can be used as the template for RNA synthesis. The region of unwinding is called a transcription bubble.
Elongation
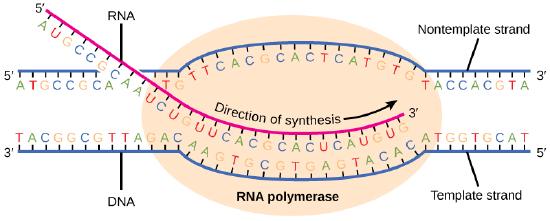
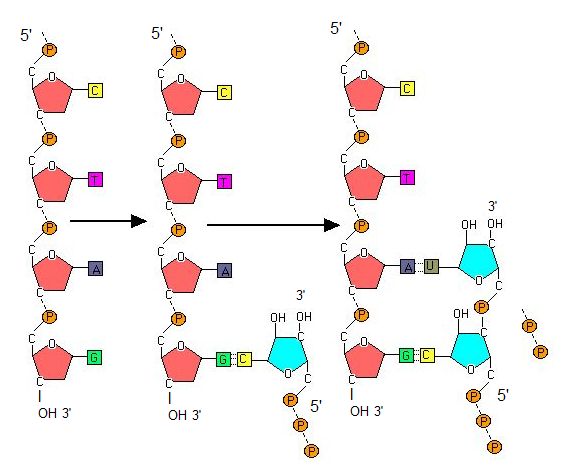
Transcription always proceeds from the template strand, one of the two strands of the double-stranded DNA. The RNA product is complementary to the template strand and is almost identical to the nontemplate strand, called the coding strand, with the exception that RNA contains a uracil (U) in place of the thymine (T) found in DNA. During elongation, an enzyme called RNA polymerase proceeds along the DNA template, adding nucleotides by base pairing with the DNA template in a manner similar to DNA replication, with the difference being an RNA strand that is synthesized does not remain bound to the DNA template. As elongation proceeds, the DNA is continuously unwound ahead of the core enzyme and rewound behind it. Note that the direction of synthesis is identical to that of synthesis in DNA—5' to 3'.
Figure 4. During elongation, RNA polymerase tracks along the DNA template, synthesizing mRNA in the 5' to 3' direction, unwinding and then rewinding the DNA as it is read.
Note: possible discussion
Compare and contrast the energy story for the addition of a nucleotide in DNA replication to the addition of a nucleotide in transcription.
Bacterial vs. eukaryotic elongation
In bacteria, elongation begins with the release of the σ subunit from the polymerase. The dissociation of σ allows the core enzyme to proceed along the DNA template, synthesizing mRNA in the 5' to 3' direction at a rate of approximately 40 nucleotides per second. As elongation proceeds, the DNA is continuously unwound ahead of the core enzyme and rewound behind it. The base pairing between DNA and RNA is not stable enough to maintain the stability of the mRNA synthesis components. Instead, the RNA polymerase acts as a stable linker between the DNA template and the nascent RNA strands to ensure that elongation is not interrupted prematurely.
In eukaryotes, following the formation of the preinitiation complex, the polymerase is released from the other transcription factors, and elongation is allowed to proceed as it does in prokaryotes with the polymerase synthesizing pre-mRNA in the 5' to 3' direction. As discussed previously, RNA polymerase II transcribes the major share of eukaryotic genes, so this section will focus on how this polymerase accomplishes elongation and termination.
Termination
In bacteria
Once a gene is transcribed, the bacterial polymerase needs to be instructed to dissociate from the DNA template and liberate the newly made mRNA. Depending on the gene being transcribed, there are two kinds of termination signals. One is protein-based and the other is RNA-based. Rho-dependent termination is controlled by the rho protein, which tracks along behind the polymerase on the growing mRNA chain. Near the end of the gene, the polymerase encounters a run of G nucleotides on the DNA template and it stalls. As a result, the rho protein collides with the polymerase. The interaction with rho releases the mRNA from the transcription bubble.
Rho-independent termination is controlled by specific sequences in the DNA template strand. As the polymerase nears the end of the gene being transcribed, it encounters a region rich in CG nucleotides. The mRNA folds back on itself, and the complementary CG nucleotides bind together. The result is a stable hairpin that causes the polymerase to stall as soon as it begins to transcribe a region rich in AT nucleotides. The complementary UA region of the mRNA transcript forms only a weak interaction with the template DNA. This, coupled with the stalled polymerase, induces enough instability for the core enzyme to break away and liberate the new mRNA transcript.
In eukaryotes
The termination of transcription is different for the different polymerases. Unlike in prokaryotes, elongation by RNA polymerase II in eukaryotes takes place 1,000–2,000 nucleotides beyond the end of the gene being transcribed. This pre-mRNA tail is subsequently removed by cleavage during mRNA processing. On the other hand, RNA polymerases I and III require termination signals. Genes transcribed by RNA polymerase I contain a specific 18-nucleotide sequence that is recognized by a termination protein. The process of termination in RNA polymerase III involves an mRNA hairpin similar to rho-independent termination of transcription in prokaryotes.
In archaea
Termination of transcription in the archaea is far less studied than in the other two domains of life and is still not well understood. While the functional details are likely to resemble mechanisms that have been seen in the other domains of life, the details are beyond the scope of this course.
Cellular location
In bacteria and archaea
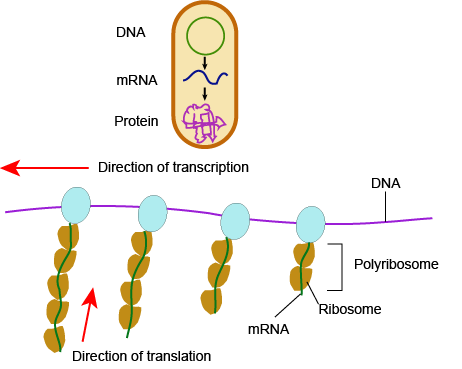
In bacteria and archaea, transcription occurs in the cytoplasm, where the DNA is located. Because the location of the DNA, and thus the process of transcription, are not physically segregated from the rest of the cell, translation often starts before transcription has finished. This means that mRNA in bacteria and archaea is used as the template for a protein before the entire mRNA is produced. The lack of spacial segregation also means that there is very little temporal segregation for these processes. Figure 6 shows the processes of transcription and translation occurring simultaneously.
In eukaryotes....
In eukaryotes, the process of transcription is physically segregated from the rest of the cell, sequestered inside of the nucleus. This results in two things: the mRNA is completed before translation can start, and there is time to "adjust" or "edit" the mRNA before translation starts. The physical separation of these processes gives eukaryotes a chance to alter the mRNA in such a way as to extend the lifespan of the mRNA or even alter the protein product that will be produced from the mRNA.
mRNA processing
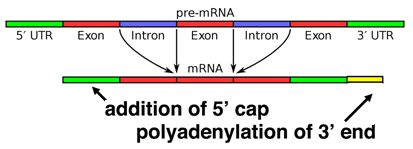
5' G-cap and 3' poly-A tail
When a eukaryotic gene is transcribed, the primary transcript is processed in the nucleus in several ways. Eukaryotic mRNAs are modified at the 3' end by the addition of a poly-A tail. This run of A residues is added by an enzyme that does not use genomic DNA as a template. Additionally, the mRNAs have a chemical modification of the 5' end, called a 5'-cap. Data suggests that these modifications both help to increase the lifespan of the mRNA (prevent its premature degradation in the cytoplasm) as well as to help the mRNA initiate translation.
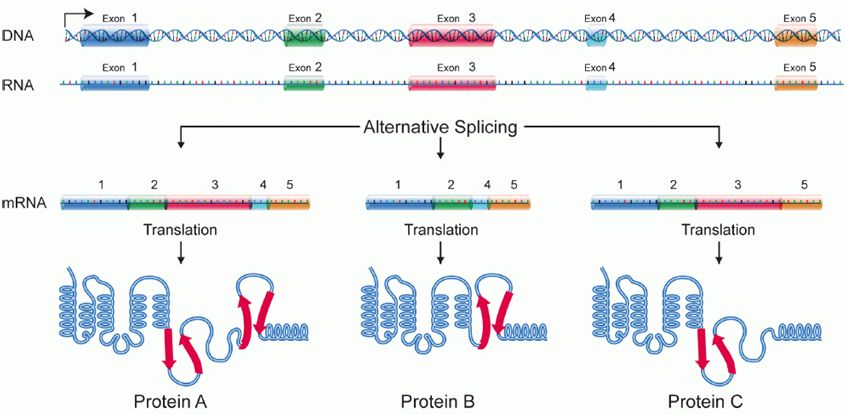
Alternative splicing
Splicing occurs on most eukaryotic mRNAs in which introns are removed from the mRNA sequence and exons are ligated together. This can create a much shorter mRNA than initially transcribed. Splicing allows cells to mix and match which exons are incorporated into the final mRNA product. As shown in the figure below, this can lead to multiple proteins being coded for by a single gene.