6.1.2: The Lux Operon
- Page ID
- 62286
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Learning Objectives
- Define quorum sensing.
- Explain the roles of regulatory proteins and autoinducers in transcriptional regulation by quorum sensing.
- Predict the effects of mutations in quorum sensing genes.
Quorum Sensing
Signaling in bacteria, known as quorum sensing, enables bacteria to monitor extracellular conditions, ensure sufficient amounts of nutrients are present, and avoid hazardous situations. There are circumstances, however, when bacteria communicate with each other.
The first evidence of bacterial communication was observed in a bacterium that has a symbiotic relationship with Hawaiian bobtail squid. When the population density of the bacteria reached a certain level, specific gene expression was initiated: the bacteria produced bioluminescent proteins that emitted light. Because the number of cells present in the environment (the cell density) is the determining factor for signaling, bacterial signaling was named quorum sensing. In politics and business, a quorum is the minimum number of members required to be present to vote on an issue.
Some species of bacteria that use quorum sensing form biofilms, which are complex colonies of bacteria (often containing several species) that exchange chemical signals to coordinate the release of toxins that attack the host. Bacterial biofilms can sometimes be found on medical equipment. When biofilms invade implants, such as hip or knee replacements or heart pacemakers, they can cause life-threatening infections.

Autoinducers in Quorum Sensing
Quorum sensing uses autoinducers as signaling molecules. Autoinducers are signaling molecules secreted by bacteria to communicate with other bacteria of the same kind. The secreted autoinducers can be small, hydrophobic molecules, such as acyl-homoserine lactone (AHL), or larger peptide-based molecules. Each type of molecule has a different mode of action. When AHL enters target bacteria, it binds to transcription factors, which then switch gene expression on or off. The peptide autoinducers stimulate more complicated signaling pathways that include bacterial kinases. The changes in bacteria following exposure to autoinducers can be quite extensive. The pathogenic bacterium Pseudomonas aeruginosa has 616 different genes that respond to autoinducers.

The Lux operon is regulated by Quorum Sensing
Bioluminesence is produced when enzymes catalyze a reaction that produces light as a by-product. The Lux operon in Vibrio fischeri includes two proteins that form an enzyme that catalyze this reaction. These bacteria live in specialized light organs in the Hawaiian bobtail squid and produce light to help the squid camouflage itself in the moonlight. However, the transcription of the genes encoding enzymes only occurs when cells are at a high density because a high concentration of autoinducer will be present.
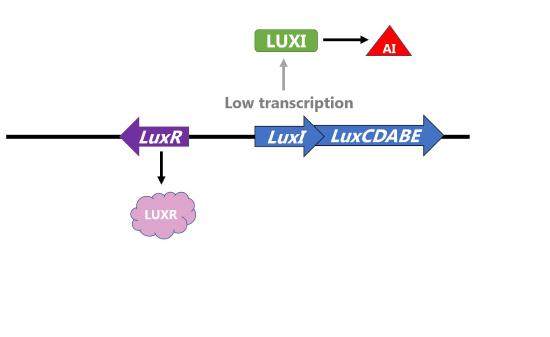
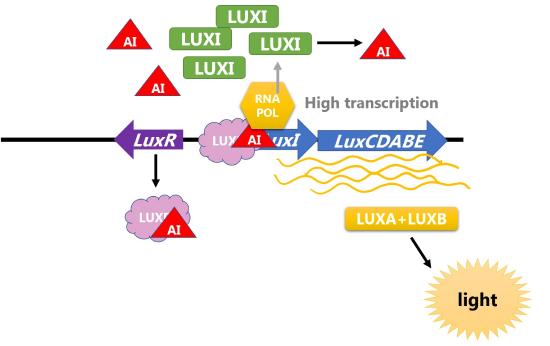
Two proteins control the amount of autoinducer and transcription of the operon. LuxI is the protein that produces the autoinducer and LuxR is the transcriptional activator that is activated in response to the autoinducer. Thus, this operon is an example of positive inducible regulation in which LuxR recruits RNA polymerase to transcribe the the operon with LuxICDABE when the concentration of bacteria (and thus autoinducer) is high. The proteins encoded by LuxA and LuxB form the luciferase enzyme that catalyzes the light-producing reaction.
Figure \(\PageIndex{3}\): Regulation of the Lux operon of V. fischeri by LUXR and bacterial density. (left) When concentration of bacteria is low, the transcription of the operon is low, levels of LUXI are low, and concentration of autoinducer (AI) is low. (right) When concentration of bacteria is high, concentration of autoinducer (AI) is high and AI binds LUXR to increase transcription of the operon. LuxA and LuxB proteins are produced and catalyze the light reaction.
Query \(\PageIndex{1}\)
Additional videos about the Lux operon and quorum sensing
Contributors and Attributions
References
Engebrecht J, Silverman M. Nucleotide sequence of the regulatory locus controlling expression of bacterial genes for bioluminescence. Nucleic Acids Res. 1987 Dec 23;15(24):10455-67. doi: 10.1093/nar/15.24.10455. PMID: 3697093; PMCID: PMC339955.