8.4: G-protein Coupled Receptors (GPCRs)
- Page ID
- 3069
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)G-protein coupled receptors are involved in responses of cells to many different kinds of signals, from epinephrine, to odors, to light. In fact, a variety of physiological phenomena including vision, taste, smell and the fight-or-flight response are mediated by GPCRs.
What are G-protein coupled receptors?
G-protein coupled receptors are cell surface receptors that pass on the signals that they receive with the help of guanine nucleotide binding proteins (a.k.a. G-proteins). Before thinking any further about the signaling pathways downstream of GPCRs, it is necessary to know a few important facts about these receptors and the G-proteins that assist them. Though there are hundreds of different G-protein coupled receptors, they all have the same basic structure: they all consist of a single polypeptide chain that threads back and forth seven times through the lipid bilayer of the plasma membrane. For this reason, they are sometimes called seven- pass transmembrane (7TM) receptors.
One end of the polypeptide forms the extracellular domain that binds the signal while the other end is in the cytosol of the cell.

When a ligand (signal) binds the extracellular domain of a GPCR, the receptor undergoes a conformational change that allows it to interact with a G-protein that will then pass the signal on to other intermediates in the signaling pathway.
What is a G-protein?
As noted above, a G-protein is a guanine nucleotide-binding protein that can interact with a G-protein linked receptor. G-proteins are associated with the cytosolic side of the plasma membrane, where they are ideally situated to interact with the cytosolic tail of the GPCR, when a signal binds to the GPCR.
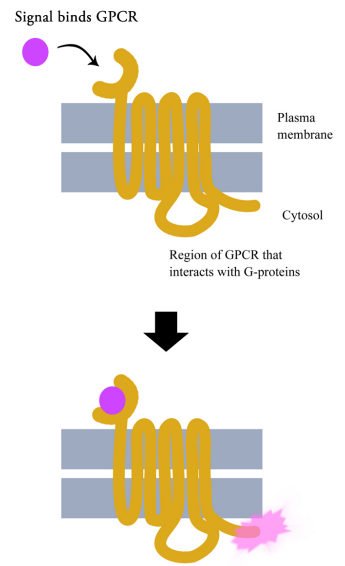
There are many different G-proteins, all of which share a characteristic structure- they are composed of three subunits called alpha, beta and gamma (aß.). Because of this, they are sometimes called heterotrimeric G proteins (hetero=different, trimeric= having three parts). The a subunit of such proteins can bind GDP or GTP and is capable of hydrolyzing a GTP molecule bound to it into GDP. In the unstimulated state of the cell, that is, in the absence of a signal bound to the GPCR, the G-proteins are found in the trimeric form (aß. bound together) and the a subunit has a GDP molecule bound to it.
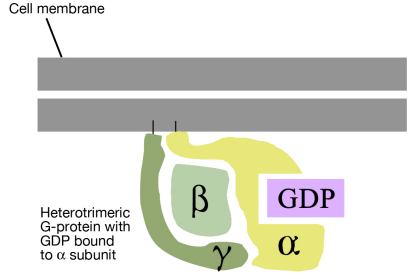
With this background on the structure and general properties of the GPCRs and the G-proteins, we can now look at what happens when a signal arrives at the cell surface and binds to a GPCR. The binding of a signal molecule by the extracellular part of the G-protein linked receptor causes the cytosolic tail of the receptor to interact with, and alter the conformation of, a G-protein. This has two consequences:
- First, the alpha subunit of the G- protein loses its GDP and binds a GTP instead.
- Second, the G-protein breaks up into the GTP-bound a part and the ß. part.
These two parts can diffuse freely along the cytosolic face of the plasma membrane and act upon their targets.

What happens when G-proteins interact with their target proteins? That depends on what the target is. G-proteins interact with different kinds of target proteins, of which we will examine two major categories:
Ion Channels
We have earlier seen that some gated ion channels can be opened or closed by the direct binding of neurotransmitters to a receptor that is an ion-channel protein. In other cases, ion channels are regulated by the binding of G-proteins. That is, instead of the signal directly binding to the ion channel, it binds to a GPCR, which activates a G-protein that then binds and opens the ion channel. The change in the distribution of ions across the plasma membrane causes a change in the membrane potential.
Specific Enzymes
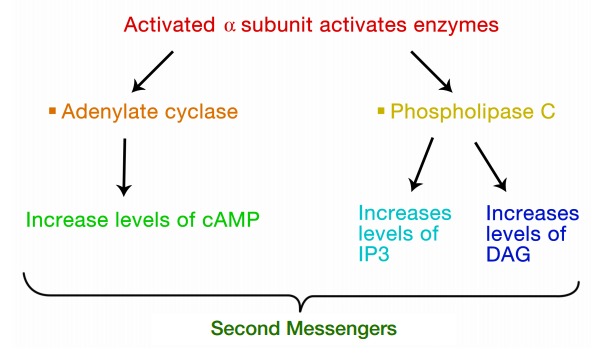
The interaction of G-proteins with their target enzymes can regulate the activity of the enzyme, either increasing or decreasing its activity. Often the target enzyme will pass the signal on in another form to another part of the cell. As you might imagine, this kind of response takes a little longer than the kind where an ion channel is opened instantaneously. Two well-studied examples of enzymes whose activity is regulated by a G-protein are adenylate cyclase and phospholipase C. When adenylate cyclase is activated, the molecule cAMP is produced in large amounts.
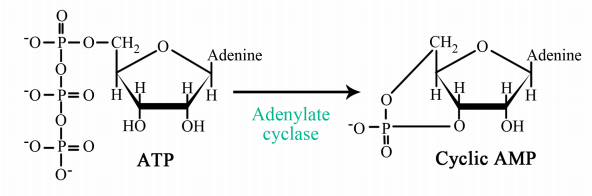
When phospholipase C is activated, the molecules inositol trisphosphate (IP3) and diacylglycerol (DAG) are made. cAMP, IP3 and DAG are second messengers, small, diffusible molecules that can "spread the message" brought by the original signal, to other parts of the cell.
In these cases, the binding of a signal to the GPCR activated a G- protein, which in turn, activated an enzyme that makes a second messenger that can amplify the message in the cell. We will first trace the effects of activating adenylate cyclase and the resulting increase in cAMP.
What is the effect of elevated cAMP levels?
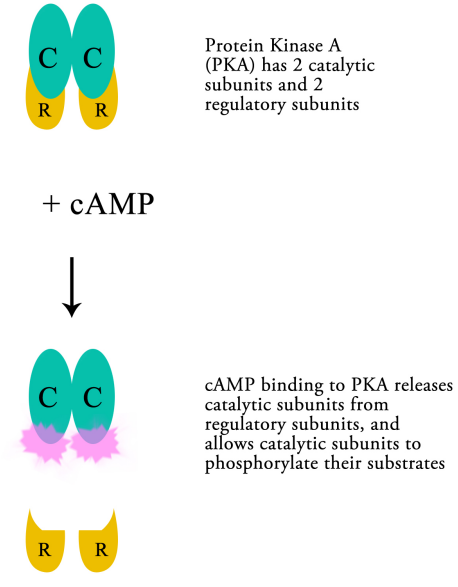
cAMP molecules bind to, and activate an enzyme, protein kinase A (PKA). PKA is composed of two catalytic and two regulatory subunits that are bound tightly together. Upon binding of cAMP the catalytic subunits are released from the regulatory subunits, allowing the enzyme to carry out its function, namely phosphorylating other proteins.

Thus, cAMP can regulate the activity of PKA, which in turn, by phosphorylating other proteins can change their activity. The targets of PKA may be enzymes that are activated by phosphorylation, or they may be proteins that regulate transcription. The phosphorylation of a transcriptional activator, for example, may cause the activator to bind to a regulatory sequence on DNA and to increase the transcription of the gene it controls. The activation of previously inactive enzymes alters the state of the cell by changing the reactions that are occurring within the cell.
For example, the binding of epinephrine to its receptor on the cell surface, activates, through the action of G-proteins, and subsequent activation of PKA, the phosphorylation of glycogen phosphorylase. The resulting activation of glycogen phosphorylase leads to the breakdown of glycogen, releasing glucose (in the form of glucose-1-phosphate) for use by the cell. Changes in gene expression, likewise, lead to changes in the cell by altering the production of particular proteins in response to the signal.

Although the steps described above seem complicated, they follow the simple pattern outlined at the beginning of this section:
- Binding of signal to receptor
- Several steps where the signal is passed on through intermediate molecules (G-proteins, adenylate cyclase, cAMP, and finally, PKA)
- Phosphorylation of target proteins by the kinase, leading to changes in the cell.
Finally, if the signal binding to the receptor serves as a switch that sets these events in motion, there must be mechanisms to turn the pathway off. The first is at the level of the G-protein. Recall that the alpha subunit of the G-protein is in its free and activated state when it has GTP bound and that it associates with the beta- gamma subunits and has a GDP bound when it is inactive. We also know that the alpha subunit has an activity that enables it to hydrolyze GTP to GDP, as shown in the figure above left. This GTP-hydrolyzing activity makes it possible for the alpha subunit, once it has completed its task, to return to its GDP bound state, re-associate with the beta-gamma part and become inactive again.

The second "off switch" is further down the signaling pathway, and controls the level of cAMP. We just noted that cAMP levels increase when adenylate cyclase is activated. When its job is done, cAMP is broken down by an enzyme called phosphodiesterase. When cAMP levels drop, PKA returns to its inactive state, putting a halt to the changes brought about by the activation of adenylate cyclase by an activated G-protein.
Let us now examine the events that follow the activation of Phospholipase C (PLC) by a G-protein. As we noted earlier, the activation of PLC results in the production of the second messengers IP3 and DAG. What do these molecules do?
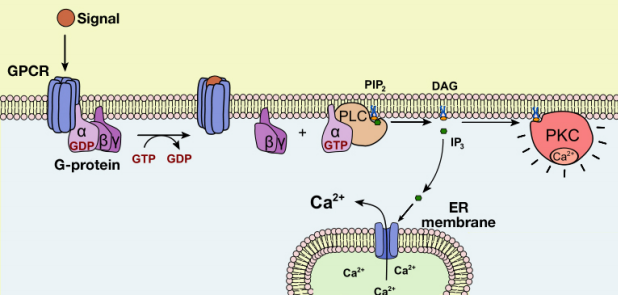
The IP3 and DAG produced by activated phospholipase C work together to activate a protein kinase. First, IP3 diffuses to the endoplasmic reticulum membrane where it binds to gated calcium ion channels. This causes calcium channels in the ER membrane to open and release large amounts of calcium into the cytoplasm from the ER lumen, as shown in the figure below.

The increase in cytosolic calcium ion concentration has various effects, one of which is to activate a protein kinase called protein kinase C (C for calcium), together with the DAG made in the earlier step. Like PKA, Protein kinase C phosphorylates a variety of proteins in the cell, altering their activity and thus changing the state of the cell.
The pathways leading to PKC and PKA activation following the binding of a signal to a GPCR are summarized in Figure 8.4.12.


