Module 4.1: Protein Structure
- Page ID
- 24722
Learning Objective
- Draw the structure of all amino acids.
- Identify chiral centers on amino acids.
- Determine the interaction of each amino acid with water.
- Join two amino acids into a dipeptide.
Amino Acids
Amino acids are the building blocks of proteins. The sequence of amino acids in individual proteins is encoded in the DNA of the cell. The physical and chemical properties of the 20 different, naturally occurring amino acids dictate the shape of the protein and its interactions with its environment. Certain short sequences of amino acids in the protein also dictate where the protein resides in the cell. Proteins are composed of hundreds to thousands of amino acids. As you can imagine, protein folding is a complicated process and there are many potential shapes due to the large number of combinations of amino acids. By understanding the properties of the amino acids you will get an appreciation for the limits of protein folding and how to predict the potential higher order structure of the protein.
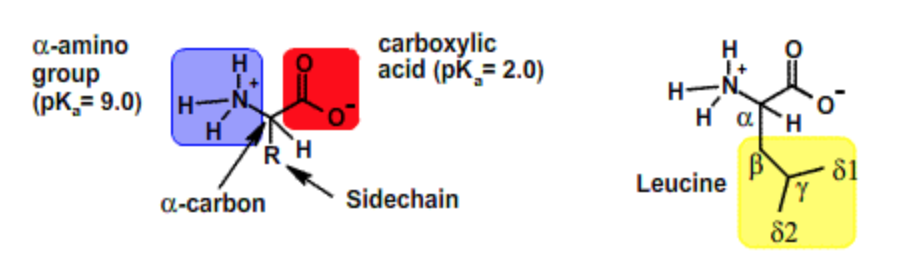
All amino acids have the same backbone structure with an amino group (the α-amino group), a carboxyl group, an α-hydrogen, and a variety of functional groups (R) all attached to the α-carbon. The atoms that are common to all amino acids are called the mainchain or backbone atoms because they will form the mainchain of the protein polymer.
The R groups, which differ from one amino acid to the next, will form the sidechain groups, because those atoms will project out to the side of the linear protein polymer.
Chirality
Because there are four different groups attached to the central carbon, the alpha carbon is an asymmetric or chiral center. This means that it is not possible to superimpose a compound on its mirror image (enantiomers).
In order to determine the absolute configuration of a chiral center follow these steps:
- Label the four groups attached to the chiral center with numbers 1-4. One(1) has the highest atomic number and four(4) has the lowest. If the atoms attached to the chiral center are the same, apply this rule to the next atom, e.g. a C-C-H would have lower priority than C-C-OH.
- Point the group labeled 4 away from you.
- If 1,2,3 is counter-clockwise the compound is S. S is latin for "sinister", or left. Imagine pointing the thumb of your left hand in the same direction as group 4, in doing so your fingers curl in the counter-clockwise direction.
- If 1,2,3 is clockwise the compound is R. R is latin for "rectus", or right. Imagine pointing the thumb of your right hand in the same direction as group 4, in doing so your fingers curl in the clockwise direction.
Although chiral compounds have the same physical properties, they generally have quite different biological properties. In the case of amino acids, and other biological compounds, the chirality of a carbon is also indicated by another labeling scheme, D and L. This scheme is based on the chirality of a reference compound, D- or L-glyceraldehyde. Bioselectivity gives rise to the dominance of one form of chirality for amino acids in nature; all common amino acids have the same chirality as L-glyceraldehyde. Although most amino acids are S, cysteine is an exception, its absolute chirality is R when the above rules are applied.
learn by doing: Determining Chirality of Iodo-fluoro-amino methane
Use the steps listed above along with the Jmol on the left below to answer the questions on the right.
1. What atom or set of atoms should be placed in the z-axis (pointing away from you) to in order to determine if the molecule is R or S?
(F = green, atomic number = 9; I = purple, atomic number = 53; N = blue, atomic number = 7; H = white, atomic number = 1)
a. F atom
b. I atom
c. H atom
d. \(NH_2\) group
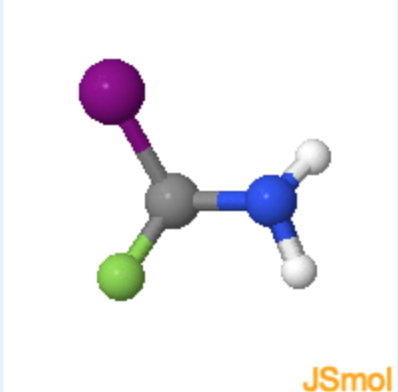
- hint
-
The substituent of lowest priority (lowest atomic number) is placed on the z-axis, or pointing away from you.
- Answer
-
c (Since H atom has the smallest molecular weight is has the least priority.)
2. Of the remaining atoms or groups on screen select their priority, one being the greatest priority 3 being the least. (F = green, 9; I = purple, 53; N = blue, 7; H = white, 1)
F atom; I atom 1/2/3; \(NH_2\) group
- hint
-
Remember that the priority increases with increased atomic number.
- Answer
-
2; 1; 3
3. Now that you know the priority of the attached atoms and groups, does this molecule have a R or S configuration? (F = green, 9; I = purple, 53; N = blue, 7; H = white, 1)
a. R
b. S
- hint
-
R is clockwise and S is counterclockwise.
- Answer
-
b. (The priority increases in a counterclockwise direction.)
learn by doing: Amino Acid Chirality
Use this Jmol to answer chirality questions.
|
L-Alanine
|
Valine
|
Cysteine
|
1. Jmol structures of valine and cysteine are shown in the middle and on the right, respectively. A static image of L-alanine is shown on the left. What is the chirality of valine, using the D, L notation?
a. D
b. L
- hint
-
rotate valine until it has the same orientation as L-alanine, with the alpha-proton pointing to the back.
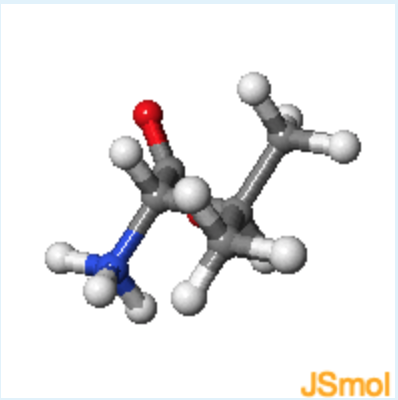
- Answer
-
b
2. Jmol structures of valine and cysteine are shown in the middle and on the right, respectively. A static image of L-alanine is shown on the left. What is the chirality of cysteine, using the D, L notation and R,S notation?
a. D and R
b. D and S
c. L and R
d. L and S
- hint
-
rotate cysteine until it has the same orientation as L-alanine, with the alpha-proton pointing to the back.
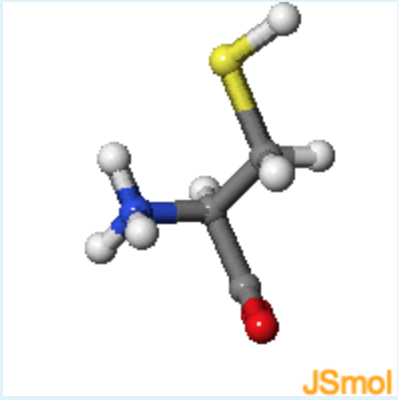
- Answer
-
c (Although the configuration is L because it is the same as L-alanine, the presence of the sulfur atom makes the beta-carbon higher in priority than the carbonyl, making the absolute configuration R instead of S.)
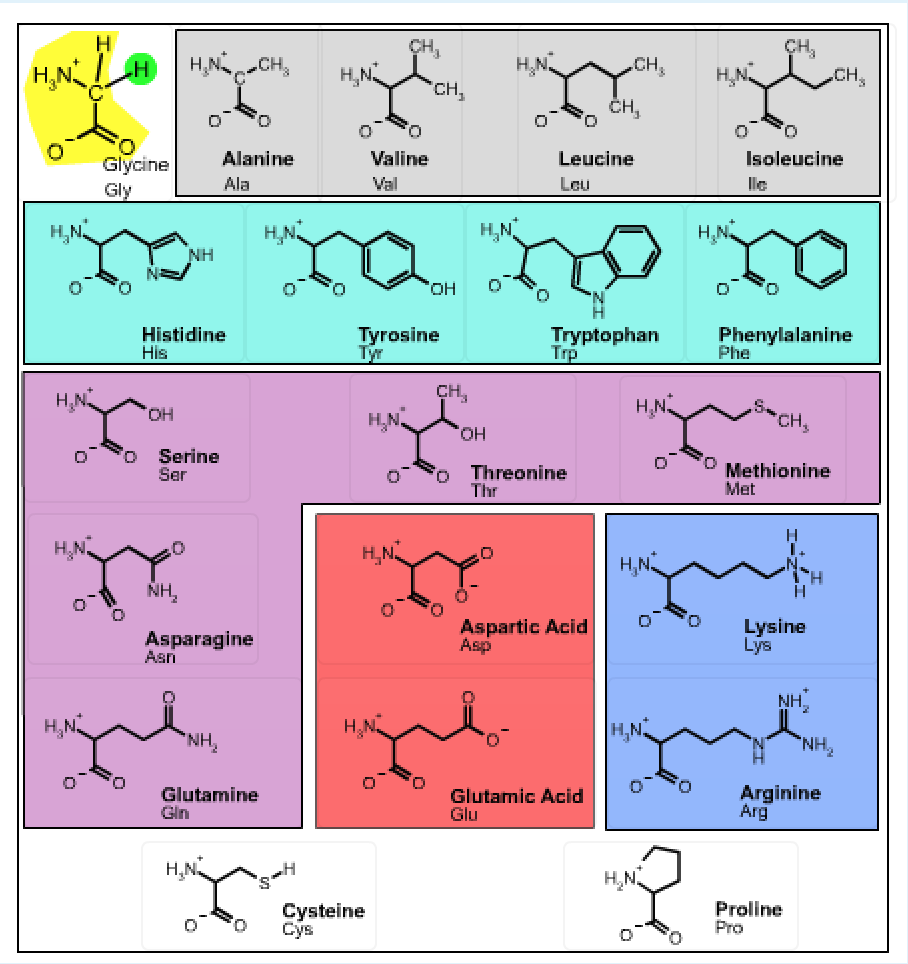
Sidechain properties: If all of the amino acids have the same basic structure with an amino, a carboxyl and a hydrogen fixed to the alpha carbon, then the large variation in the properties and structure of the amino acids must come from the fourth group attached to the alpha carbon. This group is referred to as the sidechain of the amino acid or the R group. These structures of the 20 common amino acids are shown below.
learn by doing: Common Amino Acids
Instructions: You should become familiar with the functional groups associated with the sidechain atoms of each amino acid. You should be able to infer the properties of the side chain from the 2D chemical diagram and the 3D structure. For example, which amino acids have polar sidechains? Which have planar aromatic groups? You can review the basic functional groups that were discussed in the first lecture by opening this activity.
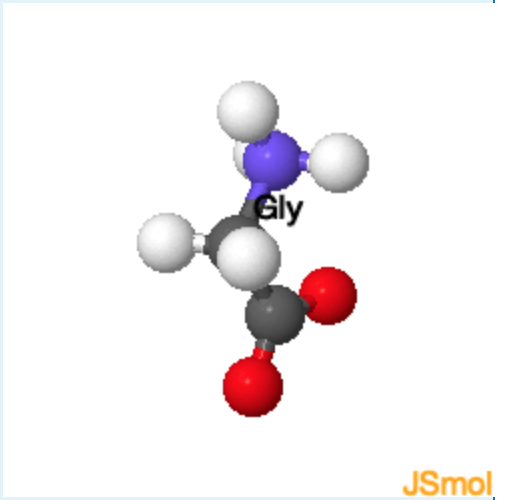
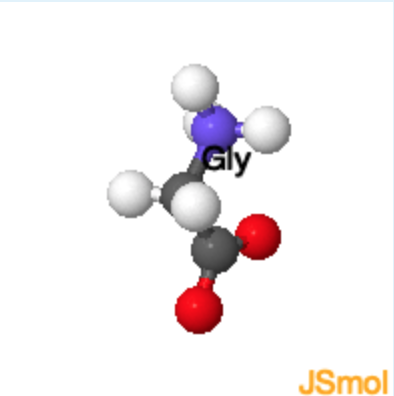
The structure of the 20 common amino acids is shown in the table. Clicking on any of the 2D drawings of an amino acid will present the 3D structure in the Jmol window on the right (Note: you may need to click twice the first time you use this tool). Initially a 2D drawing of the simplest amino acid, glycine, is shown in the upper left and its 3D structure is shown on the right.
- The mainchain atoms of glycine are highlighted in yellow and its sidechain (H) is highlighted in green. All amino acids have the same mainchain atoms, but differ in the sidechains. For clarity, the α–hydrogen is omitted in the remaining drawings.
- Non-polar amino acids are highlighted in grey,
- Aromatic amino acids are highlighted in cyan,
- Polar amino acids are highlighted in purple,
- Amino acids with acidic sidechains are highlighted in red, and
- Amino acids with basic sidechains are highlighted in blue.
- The amino acids cysteine and proline, which are shown at the bottom of the page have unique properties:
- Cysteine can form covalent S-S disulfide bond, stabilizing the protein structure by crosslinking
- Proline - the sidechain attaches to its own nitrogen, giving a secondary amine.
Acid-Base Properties of Amino Acids and Their Side-Chains
The ionization properties of amino acids depend on the mainchain amino and carboxyl group for all amino acids. Thus every amino acid has at least two ionizable groups. The sidechains of a number of amino acids have \(pK_a) values in the range of 2-12 and thus can potentially ionize in biochemical systems. The structures of these sidechains, along with their \(pK_a) values are shown in the figure below. At neutral pH (7.0) the mainchain amino group of an amino acid is positively charged and the mainchain carboxyl group is negatively charged. For those amino acids with uncharged sidechains, the positive charge on the mainchain amino group will cancel the negative charge on the carboxyl group, giving a net charge of zero, such a molecule is called a zwitterion.
The overall protonation state of an amino acid depends on the pH of the solution and the \(pK_a) values of its ionizable groups. The overall charge at any pH can be calculated using the formula:
\[q_{T o t a l}=\sum_{i=1, n} f_{H A} q_{H A}+f_{A-} q_{A-}\nonumber\]
An example of calculating the charge on the amino acid glycine is given in the previous lecture.
The isoelectric pH (pI) is the pH where a molecule (amino acid, protein, etc.) has no net charge. Using the above formula, it is possible to show that the pI for an amino acid that does not have an ionizable sidechain is the average of the \(pK_a) for the amino and carboxyl groups: pI = (1/2)(\(pK_a)NH2+\(pK_a)COOH). This simple formula does not apply to amino acids with ionizable side chains, in which case the general formula for calculating the charge (shown above) has to be used to find the pH where the average charge on the molecule is zero.
Ionizable Amino acids
The protonated (left) and deprotonated (right) forms of ionizable amino acids are shown. Note that the acidic residues become negatively charged when ionized, while the basic residues become neutral. Clicking on the image will enlarge it.
UV Absorption of Amino Acids
Tryptophan (Trp), tyrosine (Tyr), and phenylalanine (Phe) contain conjugated aromatic rings. Consequently, they absorb light in the ultraviolet range (UV). Light absorbance is quantified by the absorbance, which is equal to log(Io/I), where Io is the intensity of the incident light and I is the intensity of the light that leaves the sample. If no light is absorbed, then I=Io and A=0. As the intensity of the transmitted light decreases, A increases.
The amount of light that is absorbed by a solution of chromophores is characterized by the molar extinction coefficient. This is the absorbance that would be measured for a 1 molar solution. The extinction coefficients for Trp, Tyr, and Phe are listed below.
| Amino Acid | Extinction Coefficient \(\varepsilon(\lamda_{MAX}\)) |
|---|---|
| Trp | 5,050 \(M^{-1}cm^{-1}\) (280 nm) |
| Tyr | 1,440 \(M^{-1}cm^{-1}\) (274 nm) |
| Phe | 220 \(M^{-1}cm^{-1}\) (257 nm) |
Due to the dominance of the absorption by Trp residues, most proteins show a maximum light absorbance at a wavelength of 280 nm.
The amount of light absorbed by a solution of concentration [X] is given by the Beer-Lambert Law: \(A=\varepsilon l[X]\), where [X] is the concentration of the absorbing species, in moles/L, and \(l\) is the pathlength of the light (usually 1 cm). Given a known extinction coefficient it is possible to measure the concentration of a protein. Note that experimentally accurate absorption measurements seldom exceed ~3.0; at this value most of the light is absorbed by the sample and very little light reaches the detector in the instrument.
Calculation of molar extinction coefficients: If a protein contains a mixture of N different chromophores, the absorbance can generally be assumed to be additive. Consequently the molar extinction coefficient for the entire protein is:
\[\varepsilon=\sum_{\text {Absorbing Groups}} \varepsilon_{i} n_{i}=\varepsilon_{T r p} n_{T r p}+\varepsilon_{T y r} n_{T y r}+\varepsilon_{P h e} n_{P h e}\nonumer\]
Peptide Bond
Proteins are polymers of amino acids. The amino acids are joined together by a condensation reaction similar to that described for the formation of the glycosidic bond in polysaccharides. Each amino acid in the polymer is referred to as a residue. Individual amino acids are joined together by the attack of the nitrogen of an amino group of one amino acid on the carbonyl carbon of the carboxyl group of another to create a covalent peptide bond and yield a molecule of water as shown below.
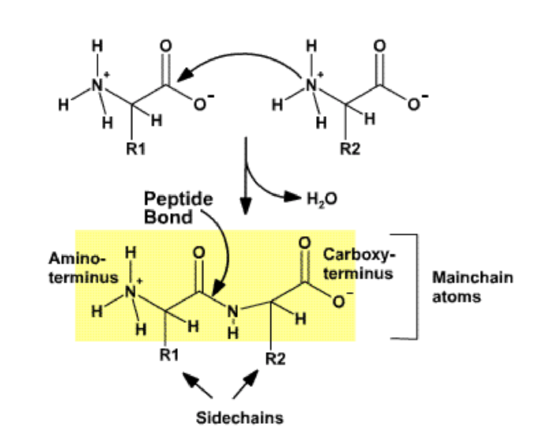
The resulting peptide chain is linear, defined by the mechanism that builds the polymer, and has defined ends. Short polymers (< 50 residues or amino acids) are usually referred to as peptides, and longer polymers as proteins. Because the synthesis takes place from the alpha amino group of one amino acid to the carboxyl group of another amino acid, the result is that there will always be a free amino group on one end of the growing polymer (the N-terminus) and a free carboxyl group on the other end (the C-terminus). Note that the potential exists for the formation of amide (peptide) links involving the carboxyl and amino groups in the side chains, but bioselectivity directs the synthesis to be linear, involving only the alpha amino and alpha carboxyl groups.
Note that after the amino acid has been incorporated into the protein, the charges on the amino and carboxy terminus have disappeared. Thus the mainchain atoms have become polar functional groups. Since each residue in a protein has exactly the same mainchain atoms, the functional properties of a protein must arise from the different sidechain groups.
By convention, the sequences of peptides and proteins are written with the N-terminus on the left and the C-terminus on the right. The name of the N-terminal residue is always the first amino acid. The name of each amino acid then follows. The primary sequence of a protein refers to its amino acid sequence.
learn by doing: Test Your Knowledge of Amino Acids
You should become familiar with the functional groups associated with the sidechain atoms of each amino acid. You should be able to infer the properties of the side chain from the 2D chemical diagram and the 3D structure. For example, which amino acids have polar sidechains? Which have planer aromatic groups? You can review the basic functional groups that were discussed in the first lecture by following this link.
 Instructions: In order to answer the following questions you will have to view the 3D structure of different amino acids in the Jmol window. You can load different structures by clicking on the name of the amino acid. Note that you may have to click twice the first time you use this tool.
Instructions: In order to answer the following questions you will have to view the 3D structure of different amino acids in the Jmol window. You can load different structures by clicking on the name of the amino acid. Note that you may have to click twice the first time you use this tool.
1. Which amino acid is this?
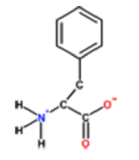
a. Histidine
b. Phenylalanine
c. Alanine
d. Tyrosine
- hint
-
You should focus on the sidechain atoms
- Answer
-
b
2. Which amino acid is aromatic (planer sidechain) and very polar?
a. Phenylalanine
b. Alanine
c. Histidine
d. Tyrosine
- hint
-
You should focus on the sidechain atoms.
- Answer
-
c (a. although planer - it is non-polar; b. although non-polar it is not aromatic; d. but close - tyrosine is aromatic and somewhat polar, but not very polar. )
3. Which amino acid has a carboxylic acid functional group on its sidechain?
a. Glutamine
b. Serine
c. Lysine
d. Glutamate
- hint
-
You should focus on the sidechain atoms.
- Answer
-
c (a. the functional group is an amide.; b. serine contains an alcohol (-OH) group; c. lysine has an amino group.)
4. Which pairs of amino acids are similar in structure, but differ in that one contains a carboxylic acid functional group and one an amide?
a. Lysine and Leucine
b. Aspartate and Asparagine
c. Aspartate and Glutamine
d. Alanine and Arginine
- hint
-
You should focus on the sidechain atoms.
- Answer
-
c (Since H atom has the smallest molecular weight is has the least priority.)
1. Which amino acid is this?

a. Histidine
b. Phenylalanine
c. Alanine
d. Tyrosine
- hint
-
The substituent of lowest priority (lowest atomic number) is placed on the z-axis, or pointing away from you.
- Answer
-
b.
5. Which pairs of amino acids both contain the same functional group, but differ in that one is more hydrophobic than the other?
a. Serine and Threonine
b. Threonine and Valine
c. Lysine and Methonine
d. Alanine and Serine
- hint
-
You should focus on the sidechain atoms.
- Answer
-
c (both of these have an alcohol functional group and threonine is more non-polar due to the additional methyl group.)
6. All of the following amino acids contain an aromatic group as part of their sidechain, except:
a. Histidine
b. Tyrosine
c. Tryptophan
d. Proline
- hint
-
Aromatic groups are planar.
- Answer
-
d. (note that the ring is non-planar and therefore not aromatic.)
Review Quiz
learn by doing
Peptides and Amino Acids
1. Which of the following is not a sensible grouping of amino acids based on their polarity properties?
a. Ala, Leu, and Val.
b. Arg, His, and Lys.
c. Phe, Trp, and Tyr.
d. Asp, Ile, and Pro.
e. Asn, Ser, and Thr.
- Answer
-
d. (a, These are all non-polar amino acids; b, These are all positively charged amino acids; c, hese are all aromatic amino acids; e. These are all polar amino acids.)
1. The molecular formula for glycine is \(C_2H_5O_2N\). What would be the molecular formula for a linear oligomer made by linking three glycine molecules together by condensation synthesis?
a. \(C_6H_{15}O_6N_3\)
b. \(C_6H_{11}O_4N_3\)
c. \(C_6H_{15}O_6N_1\)
d. \(C_6H_{19}O_8N_3\)
e. None of the above
- hint
-
Review the condensation reaction on the page.
- Answer
-
b. (two (2) waters are liberated during the condensation reactions.)
3. List one amino acid whose sidechain is uncharged at neutral pH and can form hydrogen bonds.
- hint
-
Pick an amino acid with a polar sidechain.
- Answer
-
The possible choices are Tyrosine, Tryptophan, Serine, Threonine, Asparagine, Glutamine. All other amino acids either have no hydrogen bonding capability or have a charge at pH=7.0.
Tyrosine, serine, and threonine have an -OH group that can both donate and accept a hydrogen bond.
Asparagine and glutamine have an amide group that can donate (via NH) and accept (via =O) a hydrogen bond
Tryptophan's NH group can donate a hydrogen bond.