3.1: Amino Acids and Peptides
- Page ID
- 14927
Introduction
Proteins are one of the most abundant organic molecules in living systems and have the most diverse range of functions of all macromolecules. Proteins may be structural, regulatory, contractile, or protective; they may serve in transport, storage, or membranes; or they may be toxins or enzymes. Each cell in a living system may contain thousands of different proteins, each with a unique function. Their structures, like their functions, vary greatly. They are all, however, polymers of alpha amino acids, arranged in a linear sequence and connected together by covalent bonds.
Alpha Amino Acid Structure
The major building blocks of proteins are called alpha (α) amino acids. As their name implies they contain a carboxylic acid functional group and an amine functional group. The alpha designation is used to indicate that these two functional groups are separated from one another by one carbon group. In addition to the amine and the carboxylic acid, the alpha carbon is also attached to hydrogen and one additional group that can vary in size and length. In the diagram below, this group is designated as an R-group. Within living organisms, there are 20 common amino acids used as protein building blocks. They differ from one another only at the R-group position. The fully protonated structure of an amino acid (at low pH) is shown in Figure \(\PageIndex{1}\).
The twenty common naturally-occurring amino acids each contain an alpha-carbon, an amino, carboxylic acid, and an R group (or side chain). The R group side chains may be either nonpolar, polar and uncharged, or charged, depending on the functional group, the pH, and the pKa of any ionizable group in the side chain.
Two other amino acids occasionally appear in proteins. One is selenocysteine, which is found in Arachea, eubacteria, and animals. Another is pyrrolysine, found in Arachea. Bacteria have been modified to incorporate two new amino acids, O-methyl-tyrosine, and p-aminophenylalanine. The yeast strain Saccharomyces cerevisiae has been engineered to incorporate five new unnatural amino acids (using the TAG nonsense codon and new, modified tRNA and tRNA synthetases) with keto groups that allow chemical modifications to the protein. We will concentrate only on the 20 abundant, naturally-occurring amino acids.
Figure \(\PageIndex{2}\) shows the twenty naturally occurring alpha-amino acids as they would appear internally within a protein sequence. The squiggles show that the alpha-amino and carboxyl groups are involved in peptide bonds to adjacent amino acids in the protein sequence. Students often assume that the alpha-amino and carboxyl groups within a protein sequence are free and not part of the peptide bond. This figure should help in resolving that misconception. The three-letter and one-letter abbreviations of each amino acid, as well as their typical pKa values, are also shown. It is important to memorize the three-letter and one-letter codes for the amino acids.
Amino acids form polymers through a nucleophilic attack by the amino group of an amino acid at the electrophilic carbonyl carbon of the carboxyl group of another amino acid. The carboxyl group of the amino acid must first be activated to provide a better leaving group than OH-. The resulting link between the amino acids is an amide link which biochemists call a peptide bond. In this reaction, water is released. In a reverse reaction, the peptide bond can be cleaved by water (hydrolysis). This is illustrated in Figure \(\PageIndex{3}\).
Proteins are polymers of twenty naturally occurring amino acids. In contrast, nucleic acids are polymers of just 4 different monomeric nucleotides. Both the sequence of a protein and its total length differentiate one protein from another. Just for an octapeptide, there are over 25 billion different possible arrangements of amino acids (820). Compare this to just 65536 different oligonucleotides (4 different monomeric deoxynucleotides) of 8 monomeric units, an 8mer (84). Hence the diversity of possible proteins is enormous.
When two amino acids link together to form an amide link, the resulting structure is called a dipeptide. Likewise, we can have tripeptides, tetrapeptides, and other polypeptides. At some point, when the structure is long enough, it is called a protein. The average molecular weight of proteins in yeast is about 50,000 with about 450 amino acids. The large protein might be titin with a molecular weight of about 3 million (about 30,0000 amino acids). A new class of very small proteins (30 or fewer amino acids and perhaps better named as polypeptides) called smORFs (small open reading frames) have recently been discovered to have significant biological activity. These are encoded directly in the genome and are produced by the same processes that produce regular proteins (DNA transcription and RNA translation). They are not the result of selective cleavage of a larger protein into smaller peptide fragments.
Figure \(\PageIndex{4}\) shows several ways to represent the structure of a polypeptide or protein, each showing differing amounts of information. Note that the atoms in the side chains are denoted alpha, beta, gamma, delta, epsilon ...
Characteristics of Amino Acids
The different R-groups have different characteristics based on the nature of atoms incorporated into the functional groups. There are R-groups that predominantly contain carbon and hydrogen and are very nonpolar or hydrophobic. Others contain polar uncharged functional groups such as alcohols, amides, and thiols. A few amino acids are basic (containing amine functional groups) or acidic (containing carboxylic acid functional groups). These amino acids are capable of forming full charges and can have ionic interactions. Each amino acid can be abbreviated using a three-letter and a one-letter code. Figure \(\PageIndex{5}\) shows groupings of the amino acid based on their side chain properties.
Click Here for a Downloadable Version of the Amino Acid Chart
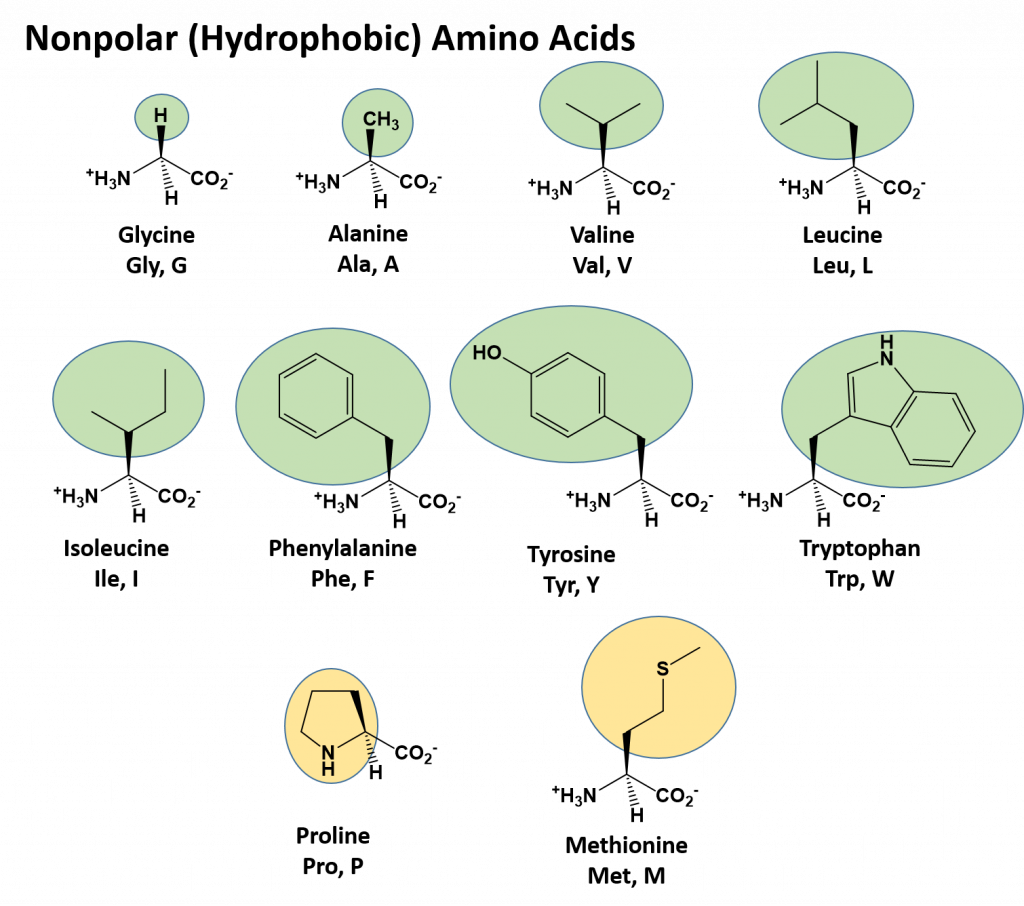
Nonpolar (Hydrophobic) Amino Acids
The nonpolar amino acids can largely be subdivided into two more specific classes, the aliphatic amino acids and the aromatic amino acids. The aliphatic amino acids (glycine, alanine, valine, leucine, isoleucine, and proline) typically contain branched hydrocarbon chains with the simplest being glycine to the more complicated structures of leucine and valine. Proline is also classified as an aliphatic amino acid but contains special properties as the hydrocarbon chain has cyclized with the terminal amine creating a unique 5-membered ring structure. As we will see in the next section covering primary structure, proline can significantly alter the 3-dimensional structure of the due to the structural rigidity of the ring structure when it is incorporated into the polypeptide chain and is commonly found in regions of the protein where folds or turns occur.
The aromatic amino acids (phenylalanine, tyrosine, and tryptophan), as their name implies, contain an aromatic functional group within their structure making them largely nonpolar and hydrophobic due to the high carbon/hydrogen content. However, it should be noted that hydrophobicity and hydrophilicity represent a sliding scale and each of the different amino acids can have different physical and chemical properties depending on their structure. For example, the hydroxyl group present in tyrosine increase its reactivity and solubility compared to that of phenylalanine.
Methionine, one of the sulfur-containing amino acids is usually classified under the nonpolar, hydrophobic amino acids as the terminal methyl group creates a thioether functional group which generally cannot form a permanent dipole within the molecule and retains low solubility.
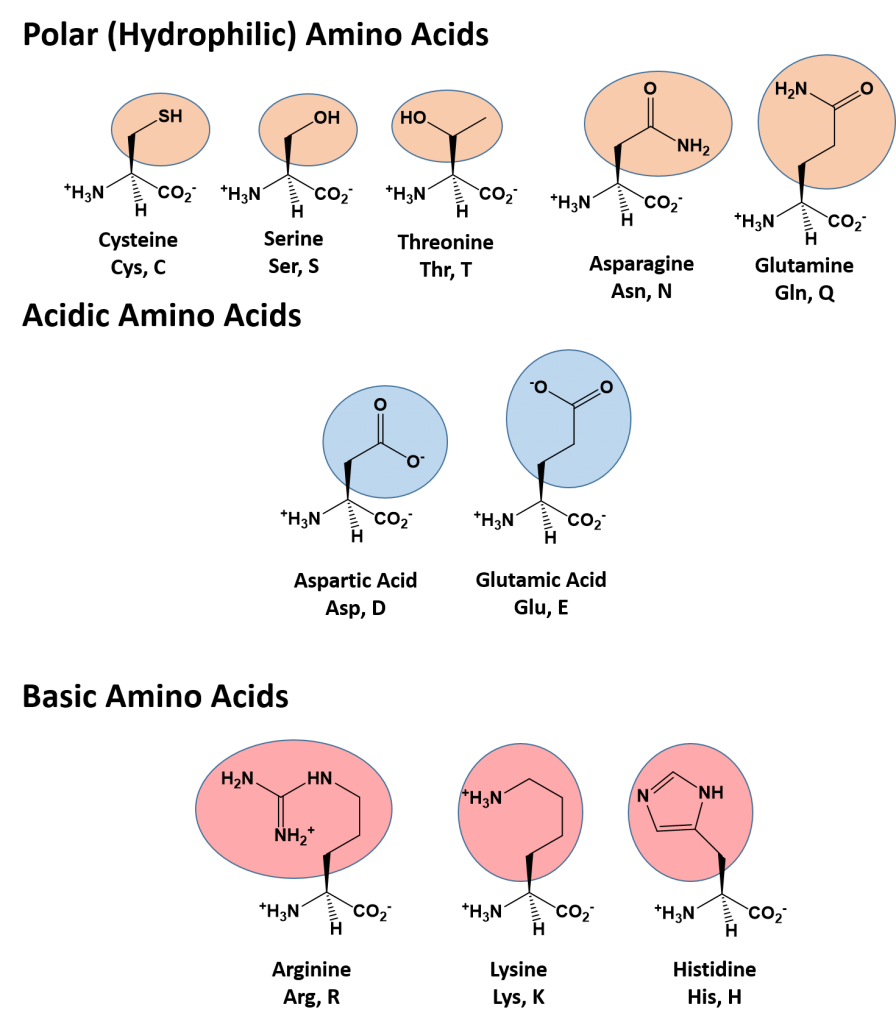
Polar (Hydrophilic) Amino Acids
The polar, hydrophilic amino acids can be subdivided into three major classes, the polar uncharged-, the acidic-, and the basic- functional groups. Within the polar uncharged class, the side chains contain heteroatoms (O, S, or N) that are capable of forming permanent dipoles within the R-group. These include the hydroxyl- and sulfhydryl-containing amino acids, serine, threonine, and cysteine, and the amide-containing amino acids, glutamine and asparagine. Two amino acids, glutamic acid (glutamate), and aspartic acid (aspartate) constitute the acidic amino acids and contain side chains with carboxylic acid functional groups capable of fully ionizing in solution. The basic amino acids, lysine, arginine, and histidine contain amine functional groups that can be protonated to carry a full charge.
Many of the amino acids with hydrophilic R-groups can participate within the active site of enzymes. An active site is the part of an enzyme that directly binds to a substrate and carries a reaction. Protein-derived enzymes contain catalytic groups consisting of amino acid R-groups that promote formation and degradation of bonds. The amino acids that play a significant role in the binding specificity of the active site are usually not adjacent to each other in the primary structure but form the active site as a result of folding in creating the tertiary structure, as you will see later in the chapter.
Thought Question: Tryptophan contains an amine functional group, why isn't tryptophan basic?
Answer: Tryptophan contains an indole ring structure that includes the amine functional group. However, due to the proximity of, and electron-withdrawing nature of the aromatic ring structure, the lone pair of electrons on the nitrogen are unavailable to accept a proton. Instead, they are involved in forming pi-bonds within several of the different resonance structures possible for the indole ring. Figure 2.3A shows four of the possible resonance structures for indole. Conversely, within the imidazole ring structure found in histidine, there are two nitrogen atoms, one of which is involved in the formation of resonance structures (Nitrogen #1 in Figure 2.3B) and cannot accept a proton, and the other (Nitrogen #3) that has a lone pair of electrons that is available to accept a proton.

Exercise \(\PageIndex{1}\)
Work It Out on Your Own:
Given the example above, describe using a chemical diagram, why the amide nitrogen atoms found in asparagine and glutamine are not basic.
- Answer
-
The lone pair is delocalized into the peptide bond (different resonance structure) so it is unavailable for sharing.
Amino Acid Stereochemistry
The amino acids are all chiral, with the exception of glycine, whose side chain is H. A chiral molecule is one that is not superimposable with its mirror image. Like left and right hands that have a thumb and fingers in the same order, but are mirror images and not the same, chiral molecules have the same things attached in the same order, but are mirror images and not the same. The mirror image versions of chiral molecules have physical properties that are nearly identical to one another, making it very difficult to tell them apart from one another or to separate them. Because of this nature, they are given a special stereoisomer name called enantiomers and in fact, the compounds themselves are given the same name! These molecules do differ in the way that they rotate plane-polarized light and the way that they react with and interact with biological molecules. Molecules that rotate light in the right-handed direction are called dextrorotary and are given a small "d" letter designation. Molecules that rotate light in the left-handed direction are called levorotary and are given a small "l" letter designation to distinguish one enantiomer from the other. Biochemists also use the older nomenclature of large "L" and "D" to characterize the 3D stereochemistry of the amino acids. All naturally occurring proteins from all living organisms consist of L amino acids, based on their structural similarities to L-glyceraldehyde.
Again, the d- and l-designations are specific terms used for the way a molecule rotates plane-polarized light. It does not denote the absolute stereo configuration of a molecule. An absolute configuration refers to the spatial arrangement of the atoms of a chiral molecular entity (or group) and its modern stereochemical description e.g. R or S, referring to Rectus, or Sinister, respectively. Absolute configurations for a chiral molecule (in pure form) are most often obtained by X-ray crystallography. Alternative techniques are optical rotatory dispersion, vibrational circular dichroism, the use of chiral shift reagents in proton NMR and Coulomb explosion imaging. When the absolute configuration is known, the assignment of R or S is based on the Cahn–Ingold–Prelog priority rules. The absolute stereochemistry is related to L-glyceraldehyde, as shown in Figure \(\PageIndex{6}\) below.
All naturally occurring amino acids in proteins are L, which corresponds to the S isomer, with the exception of cysteine. As shown in the bottom left of the Figure 6 below, the absolute configuration of the amino acids can be shown with the H pointed to the rear, the COOH groups pointing out to the left, the R group to the right, and the NH3 group upwards. You can remember this with the mnemonic "CORN".
Figure \(\PageIndex{6}\): Stereochemistry of amino acids
Why do Biochemistry still use D and L for sugars and amino acids? This explanation (taken from a website, which may not be available anymore so no reference is available) seems reasonable.
"In addition, however, chemists often need to define a configuration unambiguously in the absence of any reference compound, and for this purpose, the alternative (R,S) system is ideal, as it uses priority rules to specify configurations. These rules sometimes lead to absurd results when they are applied to biochemical molecules. For example, as we have seen, all of the common amino acids are L, because they all have exactly the same structure, including the position of the R group if we just write the R group as R. However, they do not all have the same configuration in the (R,S) system: L-cysteine is also (R)-cysteine, but all the other L-amino acids are (S), but this just reflects the human decision to give a sulfur atom a higher priority than a carbon atom and does not reflect a real difference in configuration. Worse problems can sometimes arise in substitution reactions: sometimes inversion of configuration can result in no change in the (R) or (S) prefix, and sometimes retention of configuration can result in a change of prefix.
It follows that it is not just conservatism or failure to understand the (R,S) system that causes biochemists to continue with D and L: it is just that the DL system fulfills their needs much better. As mentioned, chemists also use D and L when they are appropriate to their needs. The explanation
given above of why the (R,S) system is little used in biochemistry is thus almost the exact opposite of reality. This system is actually the only practical way of unambiguously representing the stereochemistry of complicated molecules with several asymmetric centers, but it is inconvenient with regular series of molecules like amino acids and simple sugars."
If you are told to draw the correct stereochemistry of a molecule with 1 chiral C (S isomer for example) and are given the substituents, you could do so easily following the R, S priority rules. However, how would you draw the correct isomer for the L isomer of the amino acid alanine? You couldn't do it without prior knowledge of the absolute configuration of the related molecule, L glyceraldehyde, or unless you remembered the anagram CORN. This disadvantage, however, is more than made up for by the fact that different L amino acids with the same absolute stereochemistry, might be labeled R or S, which makes this nomenclature unappealing to biochemists.
Amino Acid Charges
Monomeric amino acids have an alpha-amino group and a carboxyl group, both of which may be protonated or deprotonated, and a R group, some of which may be protonated or deprotonated. When protonated, the amino group has a +1 charge, and the carboxyl group a zero charge. When deprotonated the amino group has no charge, while the carboxyl group has a -1 charge. The R groups which can be protonated/deprotonated include Lys, Arg, and His, which have a + 1 charge when protonated, and Glu and Asp (carboxylic acids), Tyr and Ser (alcohols) and Cys (thiol), which have 0 charges when protonated. Of course, when the amino acids are linked by peptide bonds (amide link), the alpha N and the carboxyl C are in an amide link, and are not charged.
However, the amino group of the N -terminal amino acid and the carboxyl group of the C-terminal amino acid of a protein may be charged. The Henderson-Hasselbalch equation gives us a way to determine the charge state of any ionizable group knowing the pKa of the group. Write each functional group capable of being deprotonated as an acid, HA, and the deprotonated form as A. The charge of HA and A will be determined by the functional group and the Henderson-Hasselbalch equation from Chapter 2.2.
\[pH = pK_a + \log \dfrac{[A^{-}]}{HA} \nonumber \]
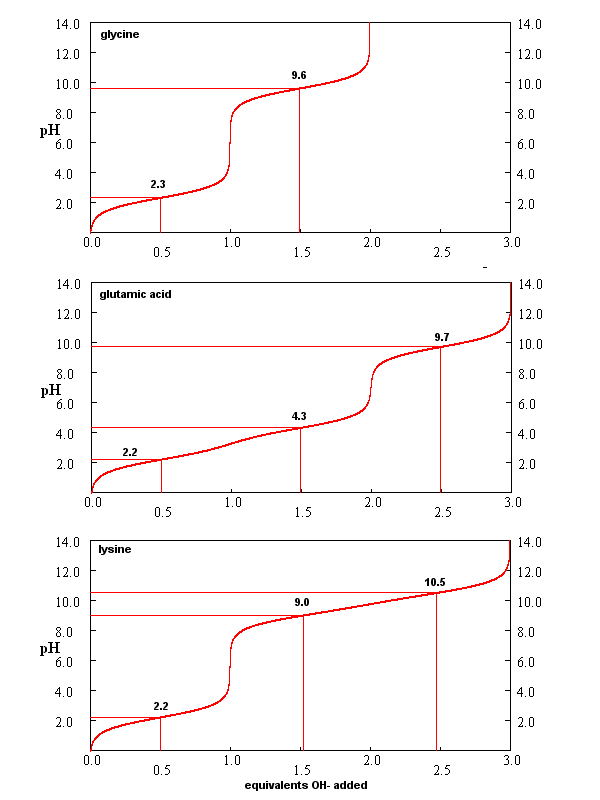
The titration curve for a single ionizable acid with different pKa values is shown below.
At the inflection point of the curve, pH = pKa and the system is most resistant to changes in pH on addition of either acid or base. At this pH, [HA]=[A-].
The properties of a protein will be determined partly by whether the side chain functional groups, the N terminal, and the C terminal are charged or not. The HH equation tells us that this will depend on the pH and the pKa of the functional group.
- If the pH is 2 units below the pKa, the HH equation becomes -2 = log A/HA, or .01 = A/HA. This means that the functional group will be about 99% protonated (with either 0 or +1 charge, depending of the functional group).
- If the pH is 2 units above the pKa, the HH equation becomes 2 = log A/HA, or 100 = A/HA. Therefore the functional group will be 99% deprotonated.
- If the pH = pKa, the HH equation becomes 0 = log A/HA, or 1 = A/HA. Therefore the functional group will be 50% deprotonated
From these simple examples, we have derived the +2 rule. This rule is used to quickly determine protonation, and hence charge state, and is extremely important to know (and easy to derive). Titration curves for Gly (no ionizable) side chain, Glu (carboxylic acid side chain) and Lys (amine side chain) are shown in Figure \(\PageIndex{7}\). You should be able to associate various sections of these curves with titration of specific ionizable groups in the amino acids.
New 5/16/23: Download this Excel spreadsheet for Titration Curves for a Triprotic Acid. It has adjustable scroll bars to change pKa values.
Buffer Review
The Henderson-Hasselbalch equation is also useful in calculating the composition of buffer solutions. Remember that buffer solutions are composed of a weak acid and its conjugate base. Consider the equilibrium for a weak acid, like acetic acid, and its conjugate base, acetate:
\[\ce{CH3CO2H + H2O <=> H3O^{+} + CH3CO2^{-}} \nonumber \]
If the buffer solution contains equal concentrations of acetic acid and acetate, the pH of the solution is:
or pH = pKa + log [A]/[HA] = 4.7 + log 1 = 4.7
A look at the titration curve for the carboxyl group of Gly (see above) shows that when the pH = pKa, the slope of the curve (i.e. the change in pH on addition of base or acid) is at a minimum. As a general rule of thumb, buffer solutions can be made for a weak acid/base in the range of +/- 1 pH unit from the pKa of the weak acids. At the pH = pKa, the buffer solution best resists the addition of either acid or base, and hence has its greatest buffering ability. The weak acid can react with the added strong base to form the weak conjugate base, and the conjugate base can react with added strong acid to form the weak acid (as shown below) so pH changes on the addition of strong acid and base are minimized.
- addition of a strong base produces a weak conjugate base: CH3CO2H + OH- ↔ CH3CO2- + H2O
- addition of strong acid produces weak acid: H3O+ + CH3CO2 → CH3CO2H + H2O
There are two simple ways to make a buffered solution. Consider an acetic acid/acetate buffer solution.
- make equal molar solution of acetic acid and sodium acetate, and mix them, monitoring pH with a pH meter, until the desired pH is reached (+/- 1 unit from the pKa).
- take a solution of acetic acid and add NaOH at substoichiometric amounts until the desired pH is reached (+/- 1 unit from the pKa). In this method you are forming the conjugate base,acetate, on the addition of the weak base:
CH3CO2H + OH- → CH3CO2- + H2O
- Buffers for pH control: Recipes based on pKas for acids, temperature, and ionic strength
Isoelectric Point
What happens if you have many ionizable groups in a single molecule, as is the case with a polypeptide or protein. Consider a protein. At a pH of 2, all ionizable groups would be protonated, and the overall charge of the protein would be positive. (Remember, when carboxylic acid side chains are protonated, their net charge is 0.) As the pH is increased, the most acidic groups will start to deprotonate and the net charge will become less positive. At high pH, all the ionizable groups will become deprotonated in the strong base, and the overall charge of the protein will be negative. At some pH, then, the net charge will be 0. This pH is called the isoelectric point (pI). The pI can be determined by averaging the pKa values of the two groups which are closest to and straddle the pI. One of the online problems will address this in more detail
Remember that pKa is really a measure of the equilibrium constant for the reaction. And of course, you remember that ΔGo = -RT ln Keq. Therefore, pKa is independent of the concentration, and depends only on the intrinsic stability of reactants with respect to the products. This is true only AT A GIVEN SET OF CONDITIONS, SUCH AS T, P, AND SOLVENT CONDITIONS.
Consider, for example, acetic acid, which in aqueous solution has a pKa of about 4.7. It is a weak acid, which dissociates only slightly to form H+ (in water the hydronium ion, H3O+, is formed) and acetate (Ac-). These ions are moderately stable in water but reassociate readily to form the starting product. The pKa of acetic acid in 80% ethanol is 6.87. This can be accounted for by the decrease in stability of the charged products which are less shielded from each other by the less polar ethanol. Ethanol has a lower dielectric constant than water. The pKa increases to 10.32 in 100% ethanol, and to a whopping 130 in air!
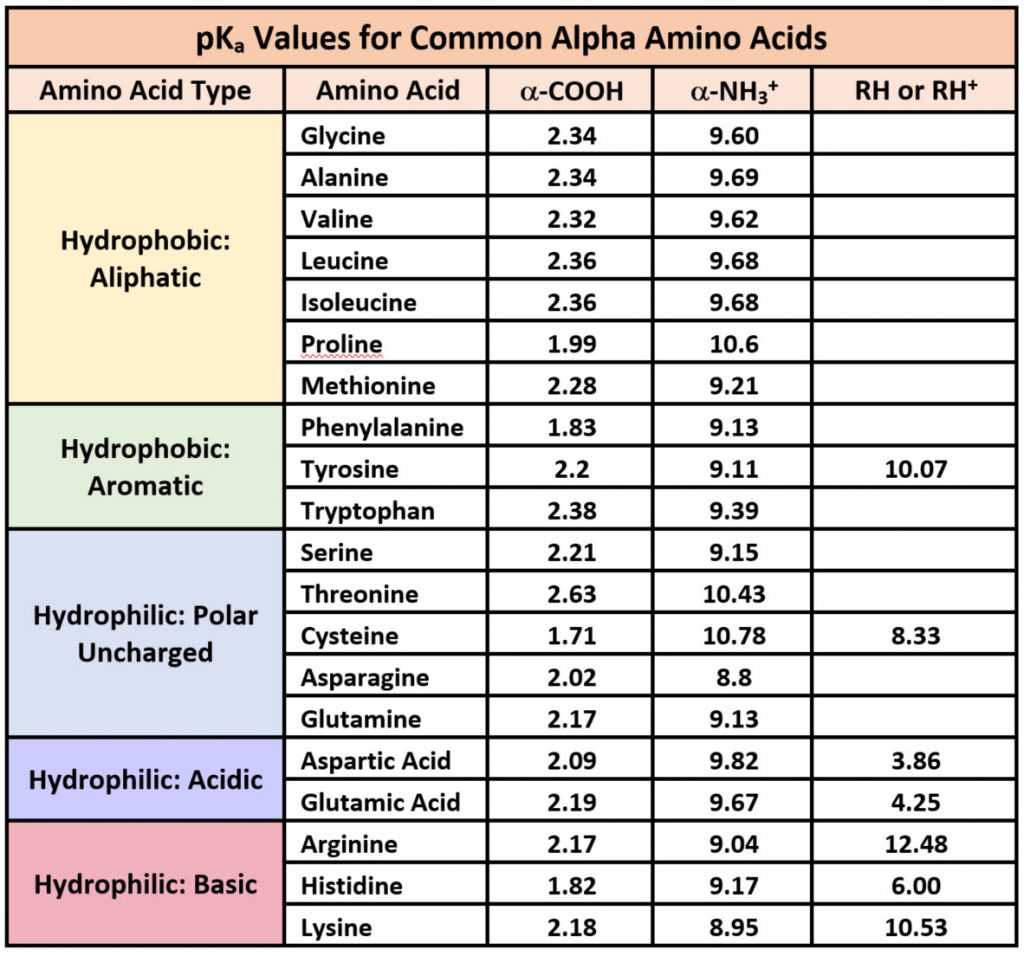
Because amino acids are zwitterions, and several also contain the potential for ionization within their R-groups, their charge state in vivo, and thus, their reactivity can vary depending on the pH, temperature, and solvation status of the local microenvironment in which they are located. Table \(\PageIndex{1}\) shows the standard pKa values for the amino acids and can be used to predict the ionization/charge status of amino acids and their resulting peptides/proteins.
Table \(\PageIndex{1}\): Summary of pKas of amino acids
However, it should be noted that the solvation status in the microenvironment of an amino acid can alter the relative pKa values of these functional groups and provide unique reactive properties within the active sites of enzymes. A more in-depth discussion of the effects of desolvation will be given in Chapter 6 discussing enzyme reaction mechanisms.
Printable Version of pKa Values
- A great interactive web site: Amino Acid Acid/Base Titration Curves
- pI calculator for any protein sequence
- Amino Acid Repository: Properties of Amino Acids
Introduction to Amino Acid Reactivity
You should be able to identify which side chains contain H bond donors and acceptors. Likewise, some are acids and bases. You should be familiar with the approximate pKa's of the side chains, and the N and C terminal groups. Three of the amino acid side chains (Trp, Tyr, and Phe) contribute significantly to the UV absorption of a protein at 280 nm. This section will deal predominantly with the chemical reactivity of the side chains, which is important in understanding the properties of the proteins. Many of the side chains are nucleophiles. Nucleophilicity is a measure of how rapidly molecules with lone pairs of electrons can react in nucleophilic substitution reactions. It correlates with basicity, which measures the extent to which a molecule with lone pairs can react with an acid (Bronsted or Lewis). The properties of the atom which holds the lone pair are important in determining both nucleophilicity and basicity. In both cases, the atom must be willing to share its unbonded electron pair. If the atoms holding the nonbonded pair is more electronegative, it will be less likely to share electrons, and that molecule will be a poorer nucleophile (nu:) and weaker base. Using these ideas, it should be clear that RNH2 is a better nucleophile than ROH, OH- is better than H2O and RSH is better than H2O. In the latter case, S is bigger and its electron cloud is more polarizable - hence it is more reactive. The important side chain nucleophiles (in order from most to least nucleophilic) are Cys (RSH, pKa 8.5-9.5), His (pKa 6-7), Lys (pKa 10.5) and Ser (ROH, pKa 13). The side chain of serine is generally no more reactive than ethanol. It is a potent nucleophile in a certain class of proteins (proteases, for example) when it is deprotonated. The amino group of lysine is a potent nucleophile only when deprotonated.
An understanding of the chemical reactivity of the various R group side chains of the amino acids in a protein is important since chemical reagents that react specifically with a given amino acid side chain can be used to:
- identify the presence of the amino acids in unknown proteins or
- determine if a given amino acid is critical for the structure or function of the protein. For example, if a reagent that covalently interacts with only Lys is found to inhibit the function of the protein, a lysine might be considered to be important in the catalytic activity of the protein.
Figure \(\PageIndex{8}\) shows a summary of nucleophilic addition and substitution at carbonyl carbons.
The rest of the section will summarize the chemistry of the side chains of reactive amino acids. Historically the function of a given amino acid in a protein has been studied by reacting them with side chain-specific chemical modifying agents. In addition, some side chains are covalently modified after they are synthesized in vivo (post-translational modification - see below).
Reactions of Lysine
Figure \(\PageIndex{9}\) the reaction of lysine with anhydrides and ethylacetimidate.
- reacts with anhydrides in a nucleophilic substitution reaction (acylation).
- reacts reversibly with methylmaleic anhydride (also called citraconic anhydride) in a nucleophilic substitution reaction.
- reacts with high specificity and yield toward ethylacetimidate in a nucleophilic substitution reaction (ethylacetimidate is like ethylacetate only with a imido group replacing the carbonyl oxygen). Ethanol leaves as the amidino group forms. (has two N -i.e. din - attached to the C)
Figure \(\PageIndex{10}\) a second set of common reactions of lysine, including those used to attach a chromophore or a fluorescent label to the side chain.
- reacts with O-methylisourea in a nucleophilic substitution reaction. with the expulsion of methanol to form a guanidino group (has 3 N attached to C, nidi)
- reacts with fluorodintirobenzene (FDNB or Sanger's reagent) or trinitrobenzenesulfonate (TNBS, as we saw with the reaction with phosphatidylethanolamine) in a nucleophilic aromatic substitution reaction to form 2,4-DNP-lysine or TNB-lysine.
- reacts with Dimethylaminonapthelenesulfonylchloride (Dansyl Chloride) in a nucleophilic substitution reaction.
Figure \(\PageIndex{11}\) shows a final common reaction we will encounter: the formation of an imine or Schiff base on the reaction of lysine with an aldehyde or ketone.
- reacts with high specificity toward aldehydes to form imines (Schiff bases), which can be reduced with sodium borohydride or cyanoborohydride to form a secondary amine.
Reactions of Cysteine
Cysteine is a potent nucleophile, which is often linked to another Cys to form a covalent disulfide bond.
Figure \(\PageIndex{12}\) shows common reagents used in the lab to label free Cys side chains. These reagents are used to alter Cys side chains to determine if they have functional significance in a protein (such as an active nucleophile in an enzyme-catalyzed reaction.
- reacts with iodoacetic acid in an SN2 reaction, adding a carboxymethyl group to the S.
- reacts with iodoacetamide in an SN2 reaction, adding a carboxyamidomethyl group to S.
- reacts with N-ethylmaleimide in an addition reaction to the double bond
Sulfur is directly below oxygen in the periodic table and, in analogy to water, sulfur-containing amino acids are found in different redox states, as illustrated in Figure \(\PageIndex{13}\).
Cystine Chemistry
Two cysteine side chains can covalently interact in a protein to produce a disulfide (RS-SR) named cystine. Just as HOOH (hydrogen peroxide) is more oxidized than HOH (O in H2O2 has an oxidation number of 1- while the O in H2O has an oxidation number of -2) , RSSR is the oxidized form (S oxidation number -1) and RSH is the reduced form (S oxidation number -2) of thiols. Their oxidation numbers are analogous since O and S are both in Group 6 of the periodic table and both are more electronegative than C.
Cystine can react with a free sulfhydryl (RSH) in a thermodynamically non-challenging disulfide exchange reaction, which when conducted with excess free sulfhydryls results in the reduction of cystine in the protein, as shown in Figure \(\PageIndex{14}\).
Figure \(\PageIndex{14}\): Disulfide interchange and reduction of protein disulfides
This reaction is often used in the lab to quantitate the amount of free cysteine side chains in a protein using Ellman's reagent, as shown in Figure \(\PageIndex{15}\).
The 2-nitro-5-thiobennzoic acid anion leaving group absorbs at 412 nm which makes quantitation easy. Only surface and not buried free cysteines will be labeled unless the protein is unfolded to expose all the cysteines.
When a protein folds, two Cys side chains might approach each other, and form an intra-chain disulfide bond. Likewise, two Cys side chains on separate proteins might approach each other and form an inter-chain disulfide. For analysis of the protein structure, disulfides are typically cleaved, and the chains are separated before analysis. The disulfides can be cleaved by reducing agents such as beta-mercaptoethanol, dithiothreitol, tris (2-carboxyethyl) phosphine (TCEP), or by oxidizing agents like performic acid which further oxidizes the disulfide to separate cysteic acids. Three common reagents used in disulfide cleavage reactions in the lab are shown in Figure \(\PageIndex{16}\).
The reaction for beta-mercaptoethanol (BME) and performic acid are shown in Figure \(\PageIndex{17}\):below.
Figure \(\PageIndex{18}\) shows the reaction for dithiothreitol (DTT). Note that it forms a stable cyclohexane-like ring, which makes this reaction very favored thermodynamically. It does not require as much of an excess of DTT as in the reaction with BME.
The reaction with tris (2-carboxyethyl) phosphine (TCEP) is not a disulfide interchange reaction as is shown in Figure \(\PageIndex{19}\).
Cells maintain a reducing environment by the presence of many "reducing" agents, such as the tripeptide gamma-Glu-Cys-Gly (glutathione). Hence intracellular proteins usually do not contain disulfides, which are abundant in extracellular proteins (such as those found in blood), or in certain organelles such as the endoplasmic reticulum and mitochondrial intermembrane space where disulfides can be introduced.
Sulfur redox chemistry is very important biologically. As described above, the sulfur in cysteine is redox-active and hence can exist in a wide variety of states, depending on the local redox environment and the presence of oxidizing and reducing agents. A potent oxidizing agent that can be made in cells is hydrogen peroxide, which can lead to more drastic and irreversible chemical modifications to the Cys side chains. If a reactive Cys is important to protein function, then the function of the protein can be modulated (sometimes reversibly, sometimes irreversibly) with various oxidizing agents, as shown in Figure \(\PageIndex{20}\).
Reactions of Histidine
Histidine is one of the strongest bases at physiological pH's. His can exist as two tautomers, as shown in Figure \(\PageIndex{21}\). NMR studies show that in model peptides, the proton predominantly is on the ε2, N3, or tele N in the imidazole ring, as it has a pKa 0.6 units higher than δ1, N1,or pro N.
The nitrogen atom in a secondary amine might be expected to be a stronger nucleophile than a primary amine through electron release to that N in a secondary amine. Opposing this effect is the steric hindrance by the two attached Cs of the N on attach on an electrophile. However, in His, this steric effect is minimized since the 2Cs are restrained by the ring. With a pKa of about 6.5, this amino acid is one of the strongest available bases at physiological pH (7.0). Hence, it can often cross-react with many of the reagents used to modify Lys side chains. His reacts with reasonably high selectivity with diethylpyrocarbonate.
In vivo Post-Translational Modification of Amino Acids
Amino acids in naturally occurring proteins are also subjected to chemical modifications within cells. These modifications alter the properties of the amino acid that is modified, which can alter the structure and function of the protein. Most chemical modifications made to proteins within cells occur after the protein is synthesized in a process called translation. The resulting chemical changes are termed post-translational modifications. Several are shown in Figure \(\PageIndex{23}\). Note that simple acid/base reactions are included, but these are not considered examples of post-translational modifications.
There are 100s of PTMs and many are part of an elaborate system within a cell to respond to both external (hormones, neurotransmitters, nutrients, metabolites) and internal chemical signals. The PTMs (like phosphorylation, acetylation, etc.) and their removal by enzymes are part of an elaborate system of cell signaling that we will explore in great detail in Chapter 28. However, not all PTMs are benign. Examples include glycation, oxidation, citrullination, and carbonylation of protein side chains. These are often increased during periods of inflammatory stress (both acute and chronic). These latter modified proteins are degraded within the cell to short peptides that retain the chemical modification. Unfortunately, these can be recognized by the immune system as foreign, which leads to an immune response against self and autoimmune disease. One such potentially deleterious PTM is the carboxyethylation of cysteine, catalyzed by the enzyme cystathionine β-synthase as shown in Figure \(\PageIndex{24}\) below.
Figure \(\PageIndex{24}\): Carboxyethylation of cysteine
The product is very similar to the carboxymethylation of cysteine shown in Figure 12 above. The modifying reagent, 3-hydroxypropionic acids, is a metabolite released by microbes found in the gut. This modification has been shown to produce an autoimmune response in the disease ankylosing spondylitis.