1.1: Introducing the Bacterial Antibiotic Sensor Mini Project
- Page ID
- 18918
Antibiotic resistance is an emerging problem in modern medicine: 70% of bacterial strains are resistant to at least one antibiotic, making treatment of common bacterial infections increasingly difficult. As a result, in the United States more people die from bacterial infections than from HIV infection and breast cancer combined. During the course of the mini project, you will learn about this important problem while getting hands-on experience with a wide variety of techniques currently used in academic and research settings: bioinformatics, site-directed mutagenesis, nucleic acid manipulation techniques and fluorescence binding assays. All of our experiments use a harmless model organism; therefore, there is no danger of getting infected.
1.1 What are Antibiotics?
The accidental discovery by Sir Alexander Fleming that the mold Penicillium notatum could destroy colonies of Staphylococcus aureus (STAPH) led to one of the greatest breakthroughs in the war against infectious diseases – the discovery of antibiotics. Commercially available antibiotics are modified natural compounds. Antibiotic scaffolds often come from natural antibiotics that are produced by soil bacteria (tetracycline, streptomycin) or fungus (penicillin). Bacteria make antibiotics for two distinctly different reasons. Firstly, at low concentration antibiotics act as signaling molecules that regulate the homeostasis of microbial communities, and may actually stimulate cell growth. In this context antibiotics play a role in cell-to-cell communication, also known as quorum sensing, and coordinate cell growth within the bacterial community. Second, at high concentrations antibiotics are agents of microbial warfare: they are produced by one bacterial species to kill another. To date, there are 160 classes of antibiotics known; most were discovered between 1940 and 1960.
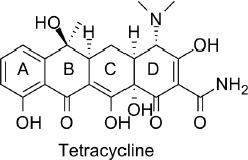
Antibiotics exert their therapeutic function by exploiting the difference in protein synthesis between bacteria and eukaryotes. The target antibiotic of the toxin sensor we study is tetracycline (Fig 1.1). Tetracycline is a polycyclic aromatic compound that halts protein synthesis in bacteria while leaving protein synthesis of the host organism unaffected. As a result tetracycline antibiotics kill bacteria without harming the patient. A commonly prescribed tetracycline derivative is doxycycline, used to treat gum disease, urinary tract infection, chlamydia and gonorrhea, among other bacterial infections.
1.2 What is Bacterial Antibiotic Resistance?
Bacterial antibiotic resistance is the ability of pathogenic bacteria to resist treatment with antimicrobial agents such as antibiotics. Antibiotic resistance is a serious threat to global health, because it jeopardizes treatment of an increasingly large number of infections caused by bacteria, fungi or a virus. According to the World Health Organization in 2012, there were about 450 000 new cases of multidrug-resistant tuberculosis. Extensively drug-resistant tuberculosis has been identified in 92 countries. Resistance to earlier generation antimalarial drugs is widespread in most malaria-endemic countries. There are high proportions of antibiotic resistance in bacteria that cause common infections (e.g. urinary tract infections, pneumonia, bloodstream infections) in all regions of the world. A high percentage of hospital-acquired infections are caused by highly resistant bacteria, such as methicillin-resistant STAPH (MRSA), or multidrug-resistant Gram-negative bacteria. Gonorrhea may soon become untreatable, because treatment failures using third-generation drugs were reported from 10 countries and no vaccines or new drugs are in development. Patients infected with a drug-resistant pathogen are at an increased risk of worse clinical outcomes (even death), and generally require more healthcare resources compared to patients infected with a non-drug resistant strain. The Center for Disease Control estimates the direct costs associated with hospital infections are as high as $45 billion dollars each year. With the emergence of bacterial strains that are resistant to multiple treatments, there is an increased urgency to understand how bacterial defense mechanisms are triggered in the presence of antibiotics.
Bacterial antibiotic resistance emerges for three reasons. (1) Antibiotics became the to-go treatment for infections and were often prescribed unnecessarily. (2) Patients sometimes do not finish their prescription, but stop taking the antibiotic once they feel better. In this case, the treatment is stopped before the infection is completely eliminated leading to propagation of resistant bacteria. (3) Bacteria have an intrinsic ability to thrive in toxic environments by becoming resistant to these toxins.
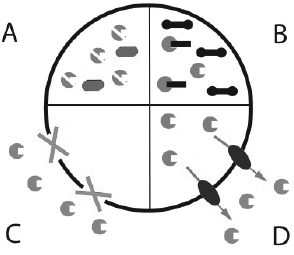
How do bacteria become resistant to antibiotics? There are four different mechanisms by which bacteria become resistant to antibiotics (Fig. 1.2). (A) The bacteria may degrade or modify the antibiotic. An example for this strategy is production of the enzyme β-lactamase that degrades antibiotics of the penicillin family. (B) Bacteria may undergo mutagenesis and change the target of the antibiotic. A good example are antibiotics that exert their function by directly binding to the ribosome thus halting bacterial translation. Once the relevant part of the ribosome is mutated, the antibiotic cannot bind to it and is no longer a successful therapeutic against that bacteria. (C) The antibiotic can be prevented from entering the cell. Mycobacterium is typically more resistant to treatment, because its waxy cell envelope prevents entry of antibiotics. (D) Antibiotics may be removed from the cell with the help of efflux pumps. Efflux pumps are integral membrane proteins that pump toxins out of the cell. They are either specific to one drug or pump multiple drugs out of the bacteria. The latter type is called multidrug-resistance efflux pump or MDR pump. The toxin sensor studied in the mini project regulates expression of a MDR efflux pump in Bacillus subtilis. Counterparts of the sensor are found in many Gram-positive bacteria.
1.3 How Do the Bacteria Detect Antibiotics In Its Environment?
Just like we do - bacteria have sensors to detect toxic compounds. Once these sensors detect an antibiotic they trigger expression of a protein that renders the antibiotic ineffective as a therapeutic using one of the four mechanisms discussed above. Toxin sensors in bacteria are usually proteins (transcription factors) that specifically bind to the toxin. The toxin-sensor protein complex then binds to the bacterial DNA or mRNA to upregulate the expression of the resistance gene that renders the antibiotic harmless to the bacteria.
In contrast, the sensor studied in the mini project is actually an RNA molecule: the ykkCD sensor RNA from Bacillus subtilis. The ykkCD sensor is encoded in the bacteria’s DNA next to a multidrug-resistant efflux pump that is also called ykkCD. The ykkCD sensor specifically recognizes the antibiotic tetracycline. The ykkCD toxin sensor binds to the antibiotic tetracycline. Tetracycline is toxic to bacteria and is a ligand of the efflux pump. Expression of the efflux pump is regulated by the ykkCD sensor. Binding of tetracycline initiates a structural change in the mRNA. This structural change enables transcription and translation of the efflux pump gene that in turns pumps tetracycline out of the bacterial cell. As a result the antibiotic tetracycline cannot be effective as therapeutic against bacteria that have the ykkCD toxin sensor or its homolog. Thus bacteria that have the ykkCD toxin sensor are resistant to tetracycline.
1.4 How Does the ykkCD Sensor Exert Its Function?
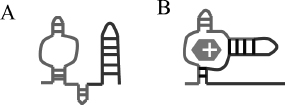
The ykkCD RNA sensor is a riboswitch. Riboswitches are conserved elements in the mRNA that regulate gene expression by allosteric structural changes. They are located in the 5’ untranslated region (5’UTR) of the gene they regulate. Most riboswitches characterized to date turn off expression of a metabolite-producing gene once sufficient amounts of metabolite is synthesized. As metabolite concentration reaches a threshold, it is able to bind to the riboswitch and initiate a structural change in the mRNA that prevents expression of the metabolite-producing gene either by halting transcription or preventing protein synthesis. The main difference between gene expression regulation by riboswitches and regulation by transcription factors is that riboswitches are part of the mRNA and are able to directly bind to their target ligand without the help of a protein cofactor. How does the ykkCD sensor undergo structural change upon tetracycline binding and directs expression of the ykkCD efflux pump? (A) In the absence of tetracycline the ykkCD sensor RNA folds into a structure that contains a terminator stem. This stem prevents synthesis of the ykkCD efflux pump mRNA. As a result the efflux pump is not made. (B) In the presence of tetracycline the ykkCD RNA sensor folds into a structure that does not contain the terminator stem. As a result the ykkCD efflux pump is made and able to pump tetracycline out of the cells (Fig. 1.3).
1.5 What Do We Do During the Mini Project?
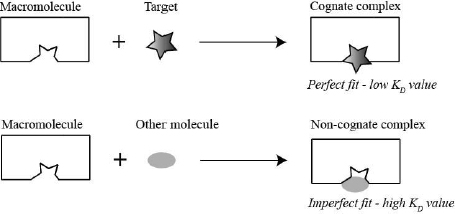
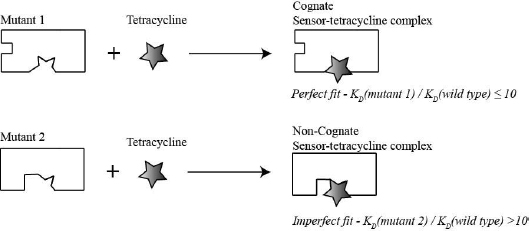
The mini project gives you a taste of hypothesis-driven real-life research and introduces you to a very important problem in biochemistry: how macromolecules recognize their specific ligand. Enzymes, transporters and receptors are all required to distinguish their specific target from other molecules. They accomplish this by forming specific noncovalent bonds (H-bonds, ion pairs and/or van der Waals interactions) with their target. The macromolecule is able to form more noncovalent interactions with their target than with other molecules therefore, the complex between the macromolecule and its target (cognate complex) is lower in energy (low dissociation constant [KD]) than the complex between the macromolecule and other molecules (non-cognate complex). Consequently, nonspecific molecules are rejected, because they cannot form all required noncovalent interactions. This selection strategy is depicted in the schematic below (Fig. 1.4).
Typically only a subset of nucleotides or amino acids in the sensor is required to recognize the specific target. How do we know which part of the macromolecule is required to recognize the target? Usually, the part of the macromolecule that is essential for recognition is evolutionary conserved. This means, it is a common research strategy to identify elements in a macromolecule that are conserved throughout evolution, change these elements by using site-directed mutagenesis and test whether the mutated macromolecule retains its ability to recognize the target ligand by performing binding assays. If the mutated macromolecule is still able to recognize the target ligand then the regions changed are most likely were not necessary to recognize the target. In contrast, if the mutated macromolecule is no longer able to recognize its target then the region changed was probably important for recognition.
In the mini project you will set out to better understand the molecular basis of how toxin sensors recognize their target. In the process you will learn state-of-the-art techniques (listed in parenthesis in the outline below). These techniques are routinely used in biochemistry and molecular biology laboratories in industry and in academia. To determine which part of the ykkCD sensor is essential for tetracycline recognition we will follow the outline below:
- (1) Identify elements within the toxin sensor that did not vary throughout evolution (sequence alignments and structure prediction).
- (2) Modify these elements using site-directed mutagenesis (primer design and PCR).
- (3) Synthesize the modified (mutated) sensors. This process involves several steps:
- Synthesis of plasmid DNA that contains the mutated sensor RNA sequence (plasmid prep)
- Enzymatic cut of this plasmid DNA (restriction digest)
- In vitro synthesis of sensor (RNA transcription)
- Purification of the mutant sensor RNA
- (4) Evaluate whether that part of the toxin sensor you choose to change was essential for tetracycline recognition or not (fluorescent binding assay). If the mutated sensor still binds strongly to tetracycline (low KD value) then the regions mutated were most likely not important for recognition. Likewise if the mutated sensor no longer binds strongly to tetracycline then the regions changed were likely to be important for recognizing tetracycline (high KD value). This thought process is illustrated on Fig. 1.5:
A flowchart of the mini project is as follows:

Note to the instructor
Chapter 1 is designed to serve as introduction to the antibiotic resistance mini project. This project was performed in eight laboratory sessions that lasted 3 hours each. It is advisable to give the introductory lecture before the start of the eight-week segment so that students are familiar with the project before they embark on it. The introductory lecture can be conveniently combined with check-in and/or safety training that often takes place when undergraduate labs meet the first time.


