19.4: DNA Replication in Prokaryotic Cells
- Page ID
- 3429
- Briefly describe the process of DNA replication.
- State why DNA can only be synthesized in a 5' to 3' direction.
- State the function of the following enzymes in bacterial DNA replication:
- DNA polymeraseIII
- DNA polymerase II
- DNA helicase
- primase
- DNA ligase
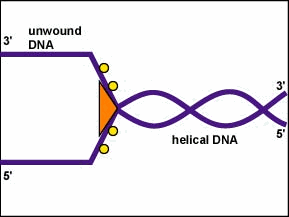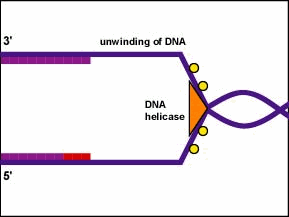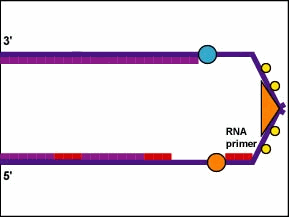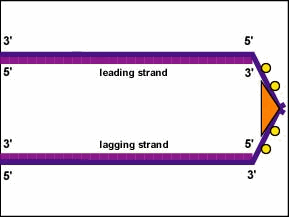
In general, DNA is replicated by uncoiling of the helix, strand separation by breaking of the hydrogen bonds between the complementary strands, and synthesis of two new strands by complementary base pairing. Replication begins at a specific site in the DNA called the origin of replication (oriC). DNA replication is bidirectional from the origin of replication. To begin DNA replication, unwinding enzymes called DNA helicases cause short segments of the two parent DNA strands to unwind and separate from one another at the origin of replication to form two "Y"-shaped replication forks. These replication forks are the actual site of DNA copying (Figure \(\PageIndex{1}\)).
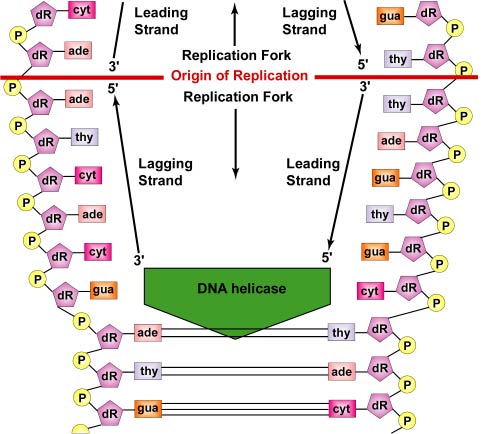
All the proteins involved in DNA replication aggregate at the replication forks to form a replication complex called a replisome (Figure \(\PageIndex{2}\)).
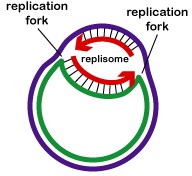
Single-strand binding proteins bind to the single-stranded regions so the two strands do not rejoin. Unwinding of the double-stranded helix generates positive supercoils ahead of the replication fork. Enzymes called topoisomerases counteract this by producing breaks in the DNA and then rejoin them to form negative supercoils in order to relieve this stress in the helical molecule during replication.
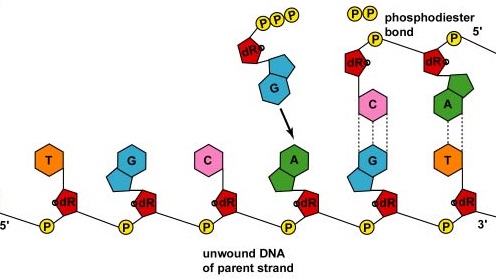
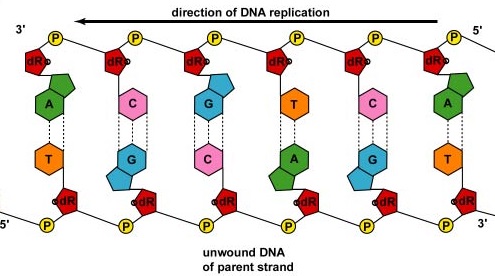
As the strands continue to unwind and separate in both directions around the entire DNA molecule, new complementary strands are produced by the hydrogen bonding of free DNA nucleotides with those on each parent strand. As the new nucleotides line up opposite each parent strand by hydrogen bonding, enzymes called DNA polymerases join the nucleotides by way of phosphodiester bonds. Actually, the nucleotides lining up by complementary base pairing are deoxynucleotide triphosphates, composed of a nitrogenous base, deoxyribose, and three phosphates. As the phosphodiester bond forms between the 5' phosphate group of the new nucleotide and the 3' OH of the last nucleotide in the DNA strand, two of the phosphates are removed providing energy for bonding (Figure \(\PageIndex{3}\)). In the end, each parent strand serves as a template to synthesize a complementary copy of itself, resulting in the formation of two identical DNA molecules (Figure \(\PageIndex{4}\)).
In bacteria, Par proteins function to separate bacterial chromosomes to opposite poles of the cell during cell division. They bind to the origin of replication of the DNA and physically pull or push the chromosomes apart, similar to the mitotic apparatus of eukaryotic cells. Fts proteins, such as FtsK in the divisome, also help in separating the replicated bacterial chromosome.
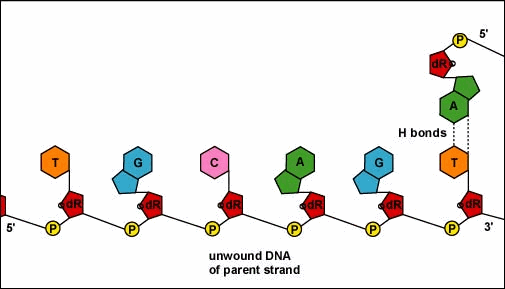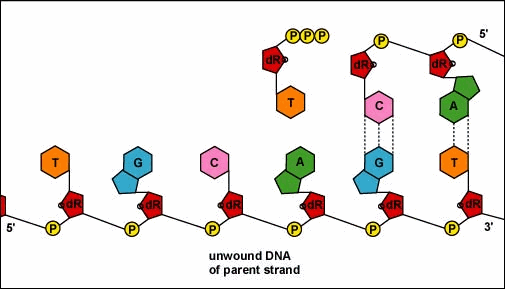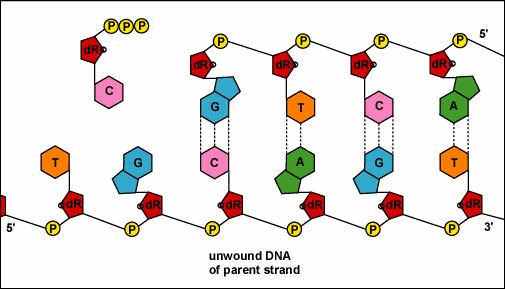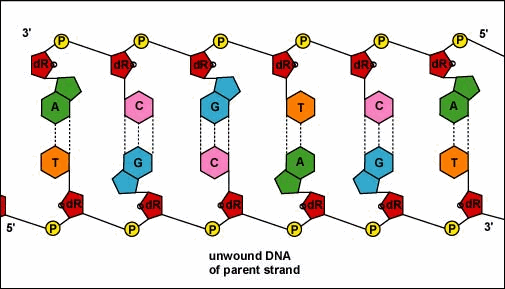
Animation: Replication of DNA by Complementary Base Pairing. As the DNA strands unwind and separate, new complementary strands are produced by the hydrogen bonding of free DNA nucleotides with those on each parent strand. As the new nucleotides line up opposite each parent strand by hydrogen bonding, enzymes called DNA polymerases join the nucleotides by way of phosphodiester bonds. The DNA polymerase responsible for these events is not shown here.
In reality, DNA replication is more complicated than this because of the nature of the DNA polmerases. DNA polymerase enzymes are only able to join the phosphate group at the 5' carbon of a new nucleotide to the hydroxyl (OH) group of the 3' carbon of a nucleotide already in the chain. As a result, DNA can only be synthesized in a 5' to 3' direction while copying a parent strand running in a 3' to 5' direction.
Remember, as mentioned above, each DNA strand has two ends. The 5' end of the DNA is the one with the terminal phosphate group on the 5' carbon of the deoxyribose; the 3' end is the one with a terminal hydroxyl (OH) group on the deoxyribose of the 3' carbon of the deoxyribose. The two strands are antiparallel, that is they run in opposite directions. Therefore, one parent strand - the one running 3' to 5' and called the leading strand - can be copied directly down its entire length (Figure \(\PageIndex{5}\)).
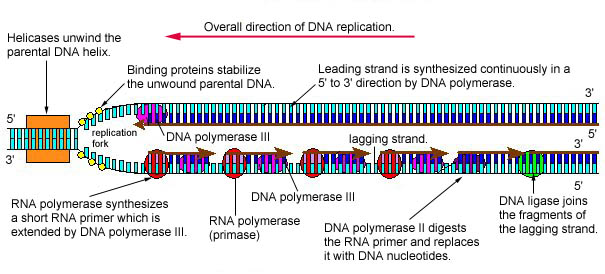
However, the other parent strand - the one running 5' to 3' and called the lagging strand - must be copied discontinuously in short fragments (Okazaki fragments) of around 100-1000 nucleotides each as the DNA unwinds. This occurs, as mentioned above, at the replisome. The lagging DNA strand loops out from the leading strand and this enables the replisome to move along both strands pulling the DNA through as replication occurs. It is the actual DNA, not the DNA polymerase that moves during bacterial DNA replication (Figure \(\PageIndex{2}\)).
In addition, DNA polymerase enzymes cannot begin a new DNA chain from scratch. They can only attach new nucleotides onto 3' OH group of a nucleotide in a preexisting strand. Therefore, to start the synthesis of the leading strand and each DNA fragment of the lagging strand, an RNA polymerase complex called a primase is required. The primase, which is capable of joining RNA nucleotides without requiring a preexisting strand of nucleic acid, first adds several complementary RNA nucleotides opposite the DNA nucleotides on the parent strand. This forms what is called an RNA primer (Figure \(\PageIndex{6}\)).
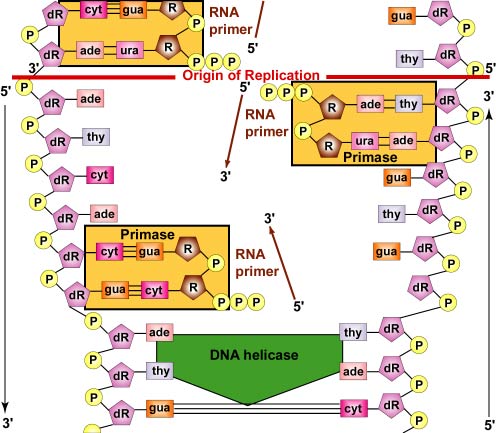
DNA polymerase III then replaces the primase and is able to add DNA nucleotides to the RNA primer (Figure \(\PageIndex{7}\)).
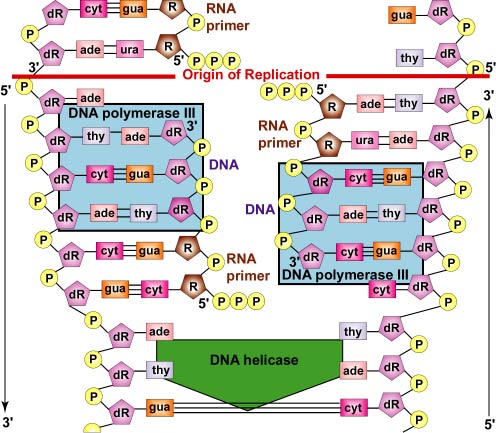
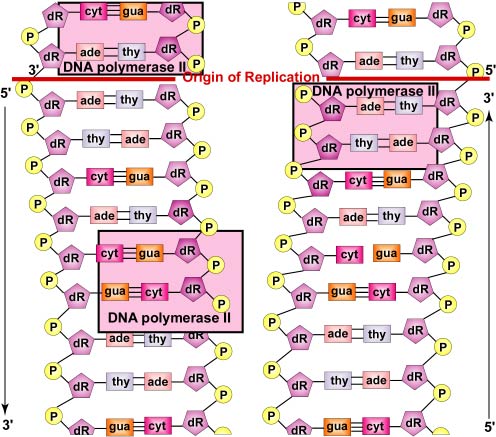
Later, DNA polymerase II digests away the RNA primer and replaces the RNA nucleotides of the primer with the proper DNA nucleotides to fill the gap (Figure \(\PageIndex{8}\)).
Animation: Replication of Leading and Lagging DNA Strands. The leading strand is made continuously in a 5' to 3' direction by DNA polymerase III as the DNA helicase unwinds the parental DNA helix. However, because the parental DNA strands are antiparallel, the lagging strand must be made in short fragments. RNA polymerase (primase) synthesizes a short RNA primer which is extended by DNA polymerase III. DNA polymerase II then digests the RNA primer and replaces it with DNA. Finally, DNA ligase joins the fragments of the lagging strand together.
There is a great deal of genetic information in the bacterial chromosome. For example Escherichia coli, the most studied of all bacteria, has a genome containing 4,639,221 base pairs, which code for at least 4288 proteins.
Summary
- During DNA replication, each parent strand acts as a template for the synthesis of the other strand by way of complementary base pairing.
- Complementary base pairing refers to DNA nucleotides with the base adenine only forming hydrogen bonds with nucleotides having the base thymine (A-T). Likewise, nucleotides with the base guanine can hydrogen bond only with nucleotides having the base cytosine (G-C).
- Each DNA strand has two ends. The 5' end of the DNA is the one with the terminal phosphate group on the 5' carbon of the deoxyribose; the 3' end is the one with a terminal hydroxyl (OH) group on the deoxyribose of the 3' carbon of the deoxyribose.
- To synthesize the two chains of deoxyribonucleotides during DNA replication, the DNA polymerase enzymes involved are only able to join the phosphate group at the 5' carbon of a new nucleotide to the hydroxyl (OH) group of the 3' carbon of a nucleotide already in the chain.
- While the two strands of DNA are complementary, they are oriented in opposite directions to each other. One strand is said to run 5' to 3'; the opposite DNA strand runs antiparallel, or 3' to 5'.
- To begin DNA replication, unwinding enzymes called DNA helicases cause short segments of the two parent DNA strands to unwind and separate from one another at the origin of replication to form two "Y"-shaped replication forks.
- Single-strand binding proteins bind to the now unpaired single-stranded regions so the two strands do not rejoin.
- As the strands continue to unwind and separate in both directions around the entire DNA molecule, new complementary strands are produced by the hydrogen bonding of free DNA nucleotides with those on each parent strand.
- As the new nucleotides line up opposite each parent strand by hydrogen bonding, enzymes called DNA polymerases join the nucleotides by way of phosphodiester bonds.
- The two strands are antiparallel, that is they run in opposite directions. Therefore, one parent strand - the one running 3' to 5' and called the leading strand- can be copied directly down its entire length. However, the other parent strand - the one running 5' to 3' and called the lagging strand- must be copied discontinuously in short fragments (Okazaki fragments) of around 100-1000 nucleotides each as the DNA unwinds.
- Furthermore, DNA polymerase enzymes cannot begin a new DNA chain from scratch. They can only attach new nucleotides onto 3' OH group of a nucleotide in a preexisting strand. Therefore, to start the synthesis of the leading strand and each DNA fragment of the lagging strand, an RNA polymerase complex called a primase is required. The primase, which is capable of joining RNA nucleotides without requiring a preexisting strand of nucleic acid, first adds several comlementary RNA nucleotides opposite the DNA nucleotides on the parent strand forming what is called an RNA primer.
- DNA polymerase III then replaces the primase and is able to add DNA nucleotides to the RNA primer. Later, DNA polymerase II digests away the RNA primer and replaces the RNA nucleotides of the primer with the proper DNA nucleotides to fill the gap.
- The DNA fragments themselves are hooked together by the enzyme DNA ligase to complete the process.


