11.3A: Pathogen-Associated Molecular Patterns (PAMPs) and Danger-Associated Molecular Patterns (DAMPs)
- Page ID
- 3274
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \) \( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)\(\newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\) \( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\) \( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\) \( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\) \( \newcommand{\Span}{\mathrm{span}}\) \(\newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\) \( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\) \( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\) \( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\) \( \newcommand{\Span}{\mathrm{span}}\)\(\newcommand{\AA}{\unicode[.8,0]{x212B}}\)
State how long it takes for early induced innate immunity to become activated and what it involves. State what is meant by pathogen-associated molecular patterns (PAMPs), and the role PAMPs play in inducing innate immunity. Name at least 5 PAMPS associated with bacteria. Name at least 2 PAMPS associated with viruses. Define DAMPs and give two examples.
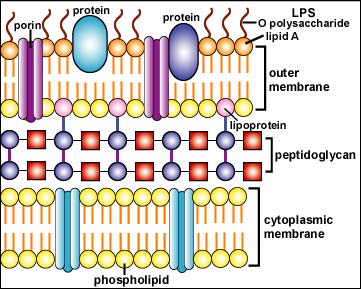
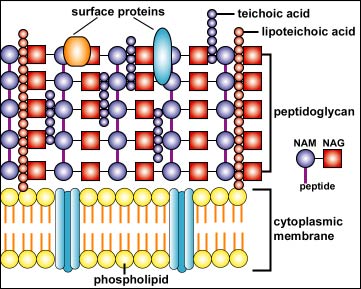
Examples of microbial-associated PAMPs include:
- lipopolysaccharide (LPS) from the outer membrane of the Gram-negative cell wall (see Figure \(\PageIndex{1}\)A);
- bacterial lipoproteins and lipopeptides (see Figure \(\PageIndex{1}\)A);
- porins in the outer membrane of the Gram-negative cell wall (see Figure \(\PageIndex{1}\)A);
- peptidoglycan found abundantly in the Gram-positive cell wall and to a lesser degree in the gram-negative cell wall (see Figure \(\PageIndex{1}\)B);
- lipoteichoic acids found in the Gram-positive cell wall (Figure \(\PageIndex{1}\)B);
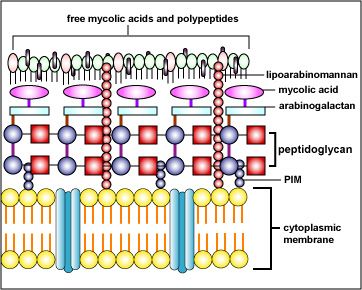
- lipoarabinomannan and mycolic acids found in acid-fast cell walls (Figure \(\PageIndex{2}\)B)
- mannose-rich glycans (short carbohydrate chains with the sugar mannose or fructose as the terminal sugar). These are common in microbial glycoproteins and glycolipids but rare in those of humans (see Figure \(\PageIndex{6}\)).
- flagellin found in bacterial flagella;
- bacterial and viral nucleic acid. Bacterial and viral genomes contain a high frequency of unmethylated cytosine-guanine dinucleotide or CpG sequences (a cytosine lacking a methyl or CH3 group and located adjacent to a guanine). Mammalian DNA has a low frequency of CpG sequences and most are methylated which may mask recognition by pattern-recognition receptors . Also, human DNA and RNA does not normally enter cellular endosomes where the pattern-recognition receptors for microbial DNA and RNA are located;
- N-formylmethionine , an amino acid common to bacterial proteins;
- double-stranded viral RNA unique to many viruses in some stage of their replication;
- single-stranded viral RNA from many` viruses having an RNA genome;
- lipoteichoic acids, glycolipids, and zymosan from yeast cell walls; and
- phosphorylcholine and other lipids common to microbial membranes.
Examples of DAMPs associated with stressed, injured, infected, or transformed host cells and not found on normal cells include:
- heat-shock proteins;
- altered membrane phospholipids; and
- molecules normally located inside phagosomes and lysosomes that enter the cytosol only when these membrane-bound compartments are damaged as a result of infection, including antibodies bound to microbes from opsonization.
- molecules normally found within cells, such as ATP, DNA, and RNA, that spill out of damaged cells.
To recognize PAMPs such as those listed above, various body cells have a variety of corresponding receptors called pattern-recognition receptors or PRRs capable of binding specifically to conserved portions of these molecules. Cells that typically have pattern recognition receptors include macrophages , dendritic cells , endothelial cells , mucosal epithelial cells, and lymphocytes .
Summary
- Early induced innate immunity begins 4 - 96 hours after exposure to an infectious agent and involves the recruitment of defense cells as a result of pathogen-associated molecular patterns or PAMPS binding to pattern-recognition receptors or PRRs.
- Pathogen-associated molecular patterns or PAMPs are molecules shared by groups of related microbes that are essential for the survival of those organisms and are not found associated with mammalian cells. Examples include LPS, porins, peptidoglycan, lipoteichoic acids, mannose-rich glycans, flagellin, bacterial and viral genomes, mycolic acid, and lipoarabinomannan.
- Danger-associated molecular patterns or DAMPs are unique molecules displayed on stressed, injured, infected, or transformed human cells also be recognized as a part of innate immunity. Examples include heat-shock proteins and altered membrane phospholipids.
- PAMPs and DAMPs bind to pattern-recognition receptors or PRRs associated with body cells to induce innate immunity.


