3.10: The Proteasome
- Page ID
- 3975
Protein degradation is as essential to the cell as protein synthesis. For example, to supply amino acids for fresh protein synthesis, to remove excess enzymes, and to remove transcription factors that are no longer needed. There are two major intracellular devices in which damaged or unneeded proteins are broken down. They are lysosomes and proteasomes
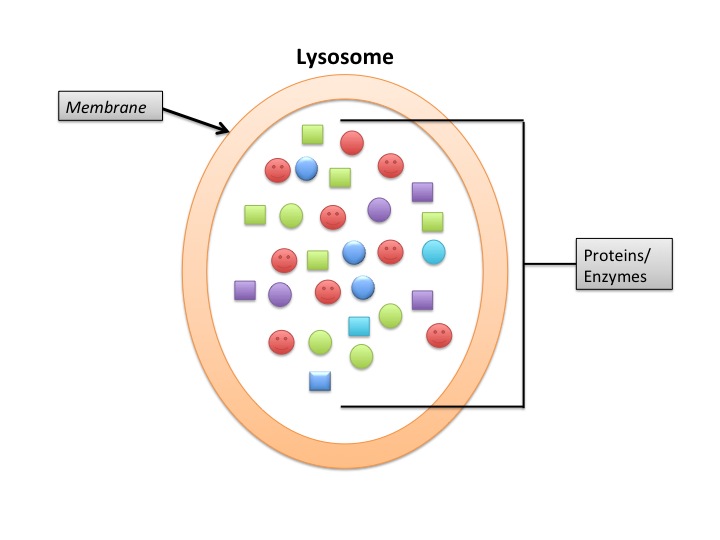
Lysosomes
Lysosomes deal primarily with extracellular proteins, e.g., plasma proteins, that are taken into the cell, e.g., by endocytosis. They are cell-surface membrane proteins that are used in receptor-mediated endocytosis. The proteins (and other macromolecules) are engulfed by autophagosomes.
Proteasomes
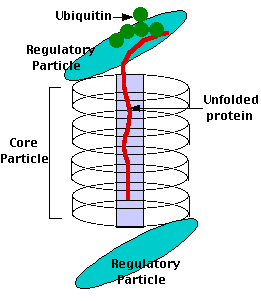
Proteasomes deal primarily with endogenous proteins; that is, proteins that were synthesized within the cell such as transcription factors, cyclins (which must be destroyed to prepare for the next step in the cell cycle) and proteins encoded by viruses and other intracellular pathogens. Proteasomes also address proteins that are folded incorrectly because of translation errors, or they are encoded by faulty genes or they have been damaged by other molecules in the cytosol. Structure of the Proteasome in the Core Particle (CP) and the Regulatory Particle (RP) as shown in Figure 3.10.2.
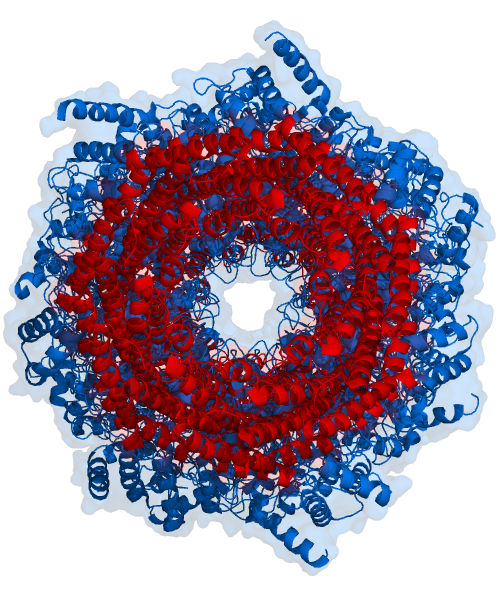
The core particle is made of 2 copies of each of 14 different proteins that are assembled in groups of 7 forming a ring. The 4 rings are stacked on each other (like 4 doughnuts) along a common center (Figure 3.10.3).

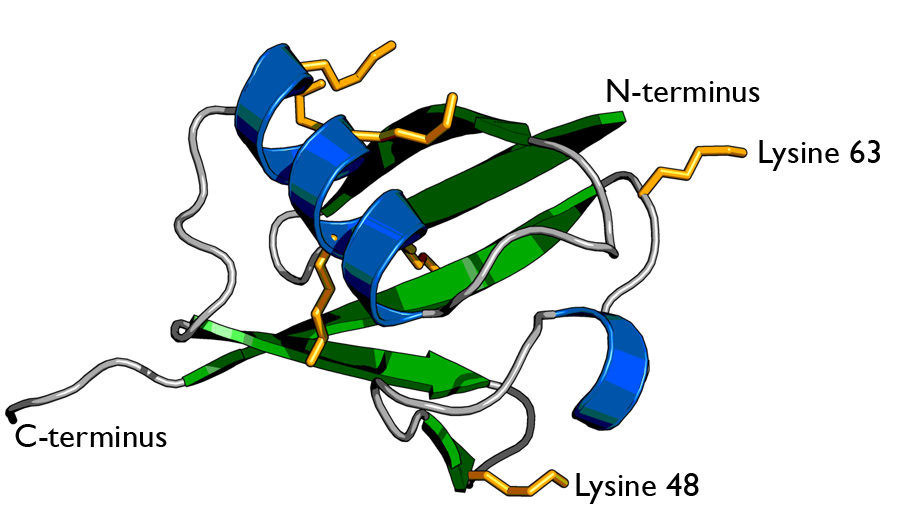
There are two identical RPs, one at each end of the core particle. Each is made of 19 different proteins (none of them the same as those in the CP). 6 of these are ATPases and some of the subunits have sites that recognize the protein ubiquitin. Ubiquitin is a small protein (76 amino acids) that is conserved throughout all the kingdoms of life (Figure 3.10.4) and is virtually identical in sequence whether in bacteria, yeast, or mammals. Ubiquitin is used by all these creatures to target proteins for destruction (hence the name based off of the "ubiquitous" term).
The Process
Proteins destined for destruction are conjugated to a molecule of ubiquitin which binds to the terminal amino group of a lysine residue. Additional molecules of ubiquitin bind to the first forming a chain and this complex then binds to ubiquitin-recognizing site(s) on the regulatory particle. The protein is unfolded by the ATPases using the energy of ATP, which is translocated into the central cavity of the core particle. Several active sites on the inner surface of the two middle "doughnuts" break various specific peptide bonds of the chain, which produces a set of peptides averaging about 8 amino acids long. These leave the core particle by an unknown route where they may be further broken down into individual amino acids by peptidases in the cytosol. However, in mammals, they may be incorporated in a class I histocompatibility molecule to be presented to the immune system as a potential antigen. The regulatory particle releases the ubiquitins for reuse
Antigen Processing by Proteasomes
In mammals, activation of the immune system leads to the release of the cytokine interferon-gamma. This causes three of the subunits in the core particle to be replaced by substitute subunits; the peptides generated in this altered proteasome are picked up by TAP (= transporter associated with antigen processing) proteins and transported from the cytosol into the endoplasmic reticulum where each enters the groove at the surface of a class I histocompatibility molecule. This complex then moves through the Golgi apparatus and is inserted in the plasma membrane where it can be "recognized" by CD8+ T cells. It is probably no coincidence that the genes encoding the three substitute core particle subunits, TAP and all the MHC (major histocompatibility complex) molecules are clustered together on the same chromosome (#6 in humans).


