5.1: DNA Replication
- Page ID
- 1646
The only way to make new cells is by the division of pre-existing cells. This means that all organisms depend on cell division for their continued existence. DNA, as you know, carries the genetic information that each cell needs. Each time a cell divides, all of its DNA must be copied faithfully so that a copy of this information can be passed on to the daughter cell. This process is called DNA replication. Before examining the actual process of DNA replication, it is useful to think about what it takes to accomplish this task successfully. Consider the challenges facing a cell in this process:
- The sheer number of nucleotides to be copied is enormous: e.g., in human cells, on the order of several billion.
- A double-helical parental DNA molecule must be unwound to expose single strands of DNA that can serve as templates for the synthesis of new DNA strands.
- This unwinding must be accomplished without introducing significant topological distortion into the molecule.
- The unwound single strands of DNA must be kept from coming back together long enough for the new strands to be synthesized.
- DNA polymerases cannot begin synthesis of a new DNA strand de novo and require a free 3' OH to which they can add DNA nucleotides.
- DNA polymerases can only extend a strand in the 5' to 3' direction. The 5' to 3' extension of both new strands at a single replication fork means that one of the strands is made in pieces.
- The use of RNA primers requires that the RNA nucleotides must be removed and replaced with DNA nucleotides and the resulting DNA fragments must be joined.
- Ensuring accuracy in the copying of so much information.
With this in mind, we can begin to examine how cells deal with each of these challenges. Our understanding of the process of DNA replication is derived from studies using bacteria, yeast, and other systems, such as Xenopus eggs. These investigations have revealed that DNA replication is carried out by the action of a large number of proteins that act together as a complex protein machine called the replisome. Numerous proteins involved in replication have been identified and characterized, including multiple different DNA polymerases in both prokaryotes and eukaryotes. Although the specific proteins involved are different in bacteria and eukaryotes, it is useful to understand the basic considerations that are relevant in all cells, before attempting to address the details of each system.
A generalized account of the steps in DNA replication is presented below, focused on the challenges mentioned above.
- The sheer number of nucleotides to be copied is enormous.
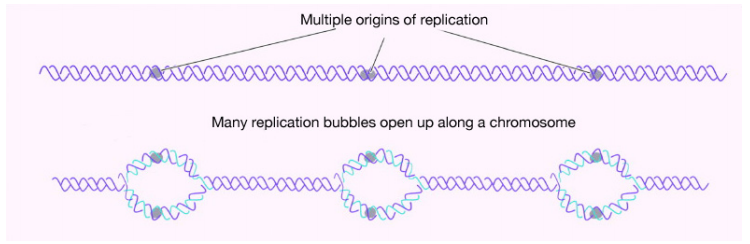
For example, in human cells, the number of nucleotides to be copied is on the order of several billion. Even in bacteria, the number is in the millions. Cells, whether bacterial or eukaryotic, have to replicate all of their DNA before they can divide. In cells like our own, the vast amount of DNA is broken up into many chromosomes, each of which is composed of a linear strand of DNA. In cells like those of E. coli, there is a single circular chromosome.
In either situation, DNA replication is initiated at sites called origins of replication. These are regions of the DNA molecule that are recognized by special origin recognition proteins that bind the DNA. The binding of these proteins helps open up a region of single-stranded DNA where the synthesis of new DNA can begin. In the case of E. coli, there is a single origin of replication on its circular chromosome. In eukaryotic cells, there may be many thousands of origins of replication, with each chromosome having hundreds. DNA replication is thus initiated at multiple points along each chromosome in eukaryotes as shown in Figure 5.1.3. Electron micrographs of replicating DNA from eukaryotic cells show many replication “bubbles" on a single chromosome.

- A double-helical parental molecule must be unwound to expose single strands of DNA that can serve as templates for the synthesis of new DNA strands.
- This unwinding must be accomplished without introducing topological distortion into the molecule.
- The unwound single strands of DNA must be kept from coming back together long enough for the new strands to be synthesized.
Once the two strands of the parental DNA molecule are separated, they must be prevented from going back together to form double-stranded DNA. To ensure that unwound regions of the parental DNA remain single-stranded and available for copying, the separated strands of the parental DNA are bound by many molecules of a protein called single-strand DNA binding protein (SSB).
- DNA polymerases cannot begin synthesis of a new DNA strand de novo and require a free 3' OH to which they can add DNA nucleotides.
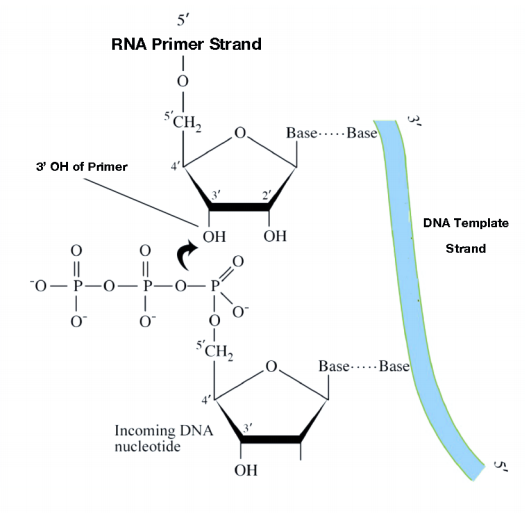
Although single-stranded parental DNA is now available for copying, DNA polymerases cannot begin synthesis of a complementary strand de novo. This is because all DNA polymerases can only add new nucleotides to the 3' end of a pre- existing chain. This means that some enzyme other than a DNA polymerase must first make a small region of nucleic acid, complementary to the parental strand, that can provide a free 3' OH to which DNA polymerase can add a deoxyribonucleotide.
This task is accomplished by an enzyme called a primase, which assembles a short stretch of RNA, called the primer, across from the parental DNA template. This provides a short base-paired region with a free 3'OH group to which DNA polymerase can add the first new DNA nucleotide (see figure on previous page). Once a primer provides a free 3'OH for extension, other proteins get into the act. These proteins are involved in loading the DNA polymerase onto the primed template and help to keep it attached to the DNA once it's on.
The first of these is the clamp loader. As its name suggests, the clamp loader helps to load a protein complex called the sliding clamp onto the DNA at the replication fork. The sliding clamp is then joined by the DNA Polymerase. The function of the sliding clamp is to increase the processivity of the DNA polymerase. This is a fancy way of saying that it keeps the polymerase associated with the replication fork by preventing it from falling off - in fact, the sliding clamp has been described as a seat-belt for the DNA polymerase.
The DNA polymerase is now poised to start synthesis of the new DNA strand (in E. coli, the primary replicative polymerase is called DNA polymerase III). As you already know, the synthesis of new DNA is accomplished by the addition of new nucleotides complementary to those on the parental strand. DNA polymerase catalyzes the reaction by which an incoming deoxyribonucleotide is added onto the 3' end of the previous nucleotide, starting with the 3'OH on the end of the RNA primer.
The 5' phosphate on each incoming nucleotide is joined by the DNA polymerase to the 3' OH on the end of the growing nucleic acid chain. As we already noted, the new DNA strands are synthesized by the addition of DNA nucleotides to the end of an RNA primer. The new DNA molecule thus has a short piece of RNA at the beginning.
- DNA polymerases can only extend a strand in the 5' to 3' direction. The 5' to 3' growth of both new strands means that one of the strands is made in pieces.
We have noted that DNA polymerase can only build a new DNA strand in the 5' to 3' direction. We also know that the two parental strands of DNA are antiparallel. This means that at each replication fork, one new strand, called the leading strand can be synthesized continuously in the 5' to 3' direction because it is being made in the same direction that the replication fork is opening up.
The synthesis of the other new strand, called the lagging strand, requires that multiple RNA primers must be laid down and the new DNA be made in many short pieces that are later joined.These short nucleic acid pieces, each composed of a small stretch of RNA primer and about 1000-2000 DNA nucleotides, are called Okazaki fragments, for Reiji Okazaki, the scientist who first demonstrated their existence.
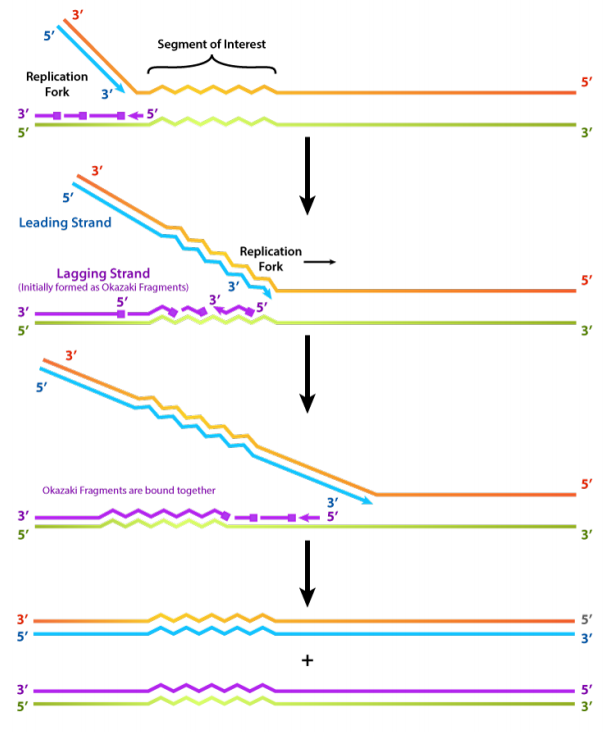
- The use of RNA primers requires that the RNA nucleotides must be removed and replaced with DNA nucleotides.
- We have seen that each newly synthesized piece of DNA starts out with an RNA primer, effectively making a new nucleic acid strand that is part RNA and part DNA. The finished DNA strand cannot be allowed to have pieces of RNA attached. o the RNA nucleotides are removed and the gaps are filled in with DNA nucleotides (by DNA polymerase I in E. coli). The DNA pieces are then joined together by the enzyme DNA ligase.
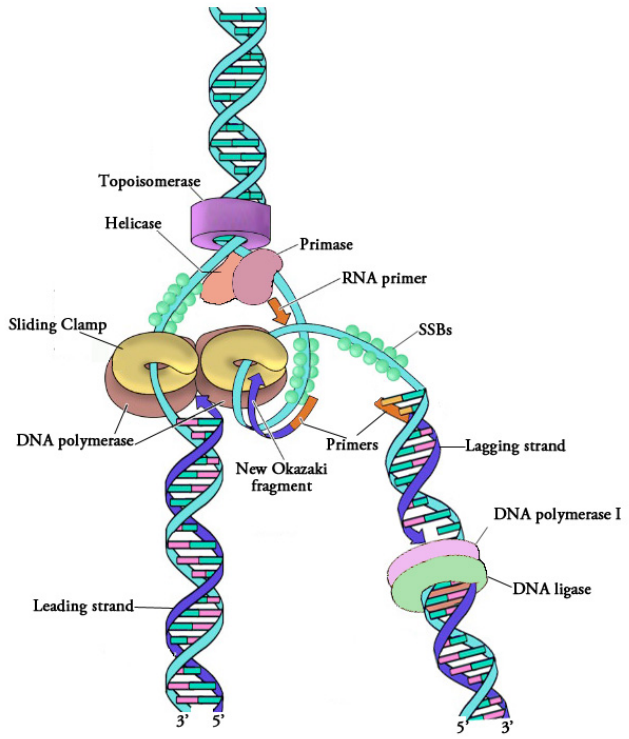
Figure 5.1.6: Proteins at a replication fork
The steps outlined above essentially complete the process of DNA replication. Figure 5.1.6 shows a replication fork, complete with the associated proteins that form the replisome.
- Ensuring accuracy in the copying of so much information
How accurate is the copying of information in the DNA by DNA polymerase? As you are aware, changes in DNA sequence (mutations) can change the amino acid sequence of the encoded proteins and that this is often, though not always, deleterious to the functioning of the organism. When billions of bases in DNA are copied during replication, how do cells ensure that the newly synthesized DNA is a faithful copy of the original information?
DNA polymerases, as we have noted earlier, work fast (averaging 50 bases a second in human cells and up to 20 times faster in E. coli). Yet, both human and bacterial cells seem to replicate their DNA quite accurately. This is because of the proof-reading function of DNA polymerases. The proof-reading function of a DNA polymerase enables the polymerase to detect when the wrong base has been inserted across from a template strand, back up and remove the mistakenly inserted base. This is possible because the polymerase is a dual-function enzyme. It can extend a DNA chain by virtue of its 5' to 3' polymerase activity but it can also backtrack and remove the last inserted base because it has a 3' to 5' exonuclease activity (an exonuclease is an enzyme that removes bases, one by one, from the ends of nucleic acids). The exonuclease activity of the DNA polymerase allows it to excise a wrongly inserted base, after which the polymerase activity inserts the correct base and proceeds with extending the strand.
In other words, DNA polymerase is monitoring its own accuracy (also termed its fidelity) as it makes new DNA, correcting mistakes immediately before moving on to add the next base. This mechanism, which operates during DNA replication, corrects many errors as they occur, reducing by about 100-fold the mistakes made when DNA is copied.


