15.6: Eukaryotic Transcription - Initiation of Transcription in Eukaryotes
- Page ID
- 13305
- Describe how transcription is initiated and proceeds along the DNA strand
Steps in Eukaryotic Transcription
Eukaryotic transcription is carried out in the nucleus of the cell by one of three RNA polymerases, depending on the RNA being transcribed, and proceeds in three sequential stages:
- Initiation
- Elongation
- Termination.
Initiation of Transcription in Eukaryotes
Unlike the prokaryotic RNA polymerase that can bind to a DNA template on its own, eukaryotes require several other proteins, called transcription factors, to first bind to the promoter region and then help recruit the appropriate polymerase. The completed assembly of transcription factors and RNA polymerase bind to the promoter, forming a transcription pre-initiation complex (PIC).
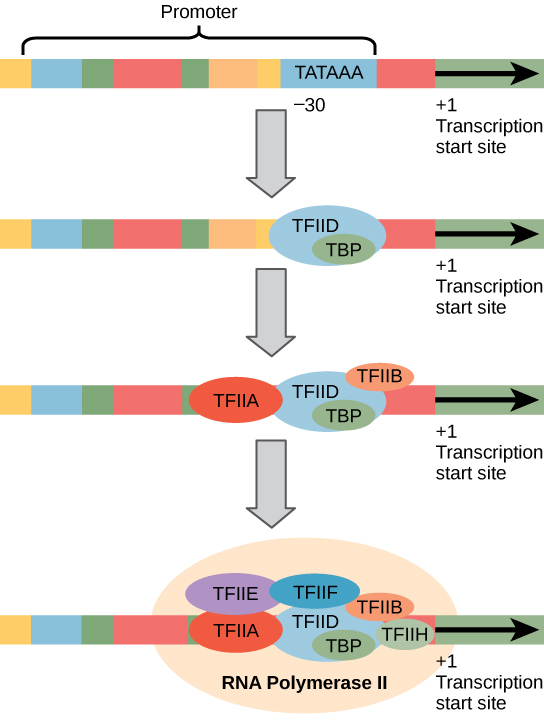
The most-extensively studied core promoter element in eukaryotes is a short DNA sequence known as a TATA box, found 25-30 base pairs upstream from the start site of transcription. Only about 10-15% of mammalian genes contain TATA boxes, while the rest contain other core promoter elements, but the mechanisms by which transcription is initiated at promoters with TATA boxes is well characterized.
The TATA box, as a core promoter element, is the binding site for a transcription factor known as TATA-binding protein (TBP), which is itself a subunit of another transcription factor: Transcription Factor II D (TFIID). After TFIID binds to the TATA box via the TBP, five more transcription factors and RNA polymerase combine around the TATA box in a series of stages to form a pre-initiation complex. One transcription factor, Transcription Factor II H (TFIIH), is involved in separating opposing strands of double-stranded DNA to provide the RNA Polymerase access to a single-stranded DNA template. However, only a low, or basal, rate of transcription is driven by the pre-initiation complex alone. Other proteins known as activators and repressors, along with any associated coactivators or corepressors, are responsible for modulating transcription rate. Activator proteins increase the transcription rate, and repressor proteins decrease the transcription rate.
The Three Eukaryotic RNA Polymerases (RNAPs)
The features of eukaryotic mRNA synthesis are markedly more complex those of prokaryotes. Instead of a single polymerase comprising five subunits, the eukaryotes have three polymerases that are each made up of 10 subunits or more. Each eukaryotic polymerase also requires a distinct set of transcription factors to bring it to the DNA template.
RNA polymerase I is located in the nucleolus, a specialized nuclear substructure in which ribosomal RNA (rRNA) is transcribed, processed, and assembled into ribosomes. The rRNA molecules are considered structural RNAs because they have a cellular role but are not translated into protein. The rRNAs are components of the ribosome and are essential to the process of translation. RNA polymerase I synthesizes all of the rRNAs except for the 5S rRNA molecule.
RNA polymerase II is located in the nucleus and synthesizes all protein-coding nuclear pre-mRNAs. Eukaryotic pre-mRNAs undergo extensive processing after transcription, but before translation. RNA polymerase II is responsible for transcribing the overwhelming majority of eukaryotic genes, including all of the protein-encoding genes which ultimately are translated into proteins and genes for several types of regulatory RNAs, including microRNAs (miRNAs) and long-coding RNAs (lncRNAs).
RNA polymerase III is also located in the nucleus. This polymerase transcribes a variety of structural RNAs that includes the 5S pre-rRNA, transfer pre-RNAs (pre-tRNAs), and small nuclear pre-RNAs. The tRNAs have a critical role in translation: they serve as the adaptor molecules between the mRNA template and the growing polypeptide chain. Small nuclear RNAs have a variety of functions, including “splicing” pre-mRNAs and regulating transcription factors. Not all miRNAs are transcribed by RNA Polymerase II, RNA Polymerase III transcribes some of them.
Modeling transcription: This interactive models the process of DNA transcription in a eukaryotic cell.
Key Points
- Eukaryotic transcription is carried out in the nucleus of the cell and proceeds in three sequential stages: initiation, elongation, and termination.
- Eukaryotes require transcription factors to first bind to the promoter region and then help recruit the appropriate polymerase.
- RNA Polymerase II is the polymerase responsible for transcribing mRNA.
Key Terms
- repressor: any protein that binds to DNA and thus regulates the expression of genes by decreasing the rate of transcription
- activator: any chemical or agent which regulates one or more genes by increasing the rate of transcription
- polymerase: any of various enzymes that catalyze the formation of polymers of DNA or RNA using an existing strand of DNA or RNA as a template


