W2017_Lecture_05_reading
- Page ID
- 7294
Amino Acid Structure
Amino acids are the monomers that make up proteins. Each amino acid has the same core structure, which consists of a central carbon atom, also known as the alpha (α) carbon, bonded to an amino group (NH2), a carboxyl group (COOH), and a hydrogen atom. Every amino acid also has another atom or group of atoms bonded to the alpha carbon known alternately as the R group, the variable group or the side-chain. For an additional introduction on amino acids, click here for a short (4 minute) video.
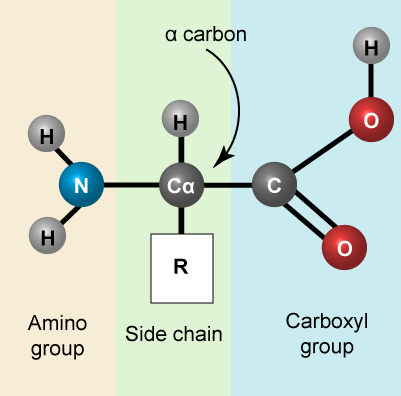
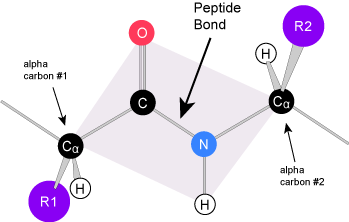
Amino acids have a central asymmetric carbon to which an amino group, a carboxyl group, a hydrogen atom, and a side chain (R group) are attached.
Attribution: Marc T. Facciotti (own work)
Possible discussion:
Recall that one of the learning goals for this class is that you (a) be able to recognize, in a molecular diagram, the backbone of an amino acid and its side chain (R-group) and (b) that you be able to draw a generic amino acid. Make sure that you practice both. You should be able to recreate something like the figure above from memory (a good use of your sketchbook is to practice drawing this structure until you can do it with the crutch of a book or the internet).
The Amino Acid Backbone
The name "amino acid" is derived from the fact that all amino acids contain both an amino group and carboxyl-acid-group in their backbone. There are 20 common amino acids present in natural proteins and each of these contain the same backbone. The backbone, when ignoring the hydrogen atoms, consists of the pattern:
N-C-C
When looking at a chain of amino acids it is always helpful to first orient yourself by finding this backbone pattern starting from the N terminus (the amino end of the first amino acid) to the C terminus (the carboxylic acid end of the last amino acid).
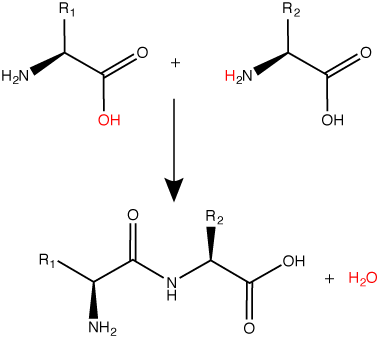
Peptide bond formation is a dehydration synthesis reaction. The carboxyl group of the first amino acid is linked to the amino group of the second incoming amino acid. In the process, a molecule of water is released and a peptide bond is formed.
Try finding the backbone in the dipeptide formed from this reaction. The pattern you are looking for is: N-C-C-N-C-C
Attribution: Marc T. Facciotti (original work)
The sequence and the number of amino acids ultimately determine the protein's shape, size, and function. Each amino acid is attached to another amino acid by a covalent bond, known as a peptide bond, which is formed by a dehydration synthesis (condensation) reaction. The carboxyl group of one amino acid and the amino group of the incoming amino acid combine, releasing a molecule of water and creating the peptide bond.
Amino Acid R group
The amino acid R group is a term that refers to the variable group on each amino acid. The amino acid backbone is identical on all amino acids, the R groups are different on all amino acids. For the structure of each amino acid refer to the figure below.

There are 20 common amino acids found in proteins, each with a different R group (variant group) that determines its chemical nature. R-groups are circled in teal. Charges are assigned assuming pH ~6.0. The full name, three letter abbreviation and single letter abbreviations are all shown.
Attribution: Marc T. Facciotti (own work)
Note:
Possible Discussion: Let's think about the relevance of having 20 different amino acids. If you were using biology to build proteins from scratch, how might it be useful if you had 10 more different amino acids at your disposal? By the way, this is actually happening in a variety of research labs - why would this be potentially useful?
Each variable group on an amino acid gives that amino acid specific chemical properties (acidic, basic, polar, or nonpolar). You should be familiar with most of the functional groups in the R groups by now. The chemical properties associated with the whole collection of individual functional groups gives each amino acid R group unique chemical potential.
For example, amino acids such as valine, methionine, and alanine are typically nonpolar or hydrophobic in nature, while amino acids such as serine and threonine are said to have polar character and possess hydrophilic side chains.
Note:
Practice: Using your knowledge of functional groups, try classifying each amino acid in the figure above as either having the tendency to be polar or nonpolar. Try to find other classification schemes and think make lists for yourself of the amino acids you would put into each group. You can also search the internet for amino acid classification schemes - you will notice that there are different ways of grouping these chemicals based on chemical properties. You may even find that there are contradictory schemes. Try to think about why this might be and apply your chemical logic to figuring out why certain classification schemes were adopted and why specific amino acids were placed in certain groups.
Protein Folding and Structure
To understand how the protein gets its final shape or conformation, we need to understand the four levels of protein structure: primary, secondary, tertiary, and quaternary. For a short (4 minutes) introduction video on protein structure click here.
Primary Structure
The unique sequence of amino acids in a polypeptide chain is its primary structure. The linear sequence of amino acids in the polypeptide chain are held together by peptide bonds and result in the N-C-C-N-C-C patterned backbone. The primary structure is coded for in the DNA, a process you will learn about in the Transcription and Translation modules.
Secondary structure
The local folding of the polypeptide in some regions gives rise to the secondary structure of the protein. The most common shapes created by secondary folding are the α-helix and β-pleated sheet structures. These secondary structures are held together by hydrogen bonds forming between the backbones of amino acids in close proximity to one another. More specifically, the oxygen atom in the carboxyl group from one amino acid can form a hydrogen bond with a hydrogen atom bound to the nitrogen in the amino group of another amino acid that is four amino acids farther along the chain.
Tertiary Structure
The unique three-dimensional structure of a polypeptide is its tertiary structure. This structure is in part due to chemical interactions at work on the polypeptide chain. Primarily, the interactions among R groups creates the complex three-dimensional tertiary structure of a protein. The nature of the R groups found in the amino acids involved can counteract the formation of the hydrogen bonds described for standard secondary structures. For example, R groups with like charges are repelled by each other and those with unlike charges are attracted to each other (ionic bonds). When protein folding takes place, the hydrophobic R groups of nonpolar amino acids lay in the interior of the protein, whereas the hydrophilic R groups lay on the outside. These types of interactions are also known as hydrophobic interactions. Interaction between cysteine side chains forms disulfide linkages in the presence of oxygen, the only covalent bond forming during protein folding.
All of these interactions, weak and strong, determine the final three-dimensional shape of the protein. When a protein loses its three-dimensional shape, it may no longer be functional.
Quaternary Structure
In nature, some proteins are formed from several polypeptides, also known as subunits, and the interaction of these subunits forms the quaternary structure. Weak interactions between the subunits help to stabilize the overall structure. For example, a multisubunit protein called insulin (a globular protein) has a combination of hydrogen bonds and disulfide bonds that hold the multiple subunits together. Each of these subunits went through primary, secondary and tertiary folding independently of one another.
Denaturation and Protein Folding
Each protein has its own unique sequence and shape that are held together by chemical interactions. If the protein is subject to changes in temperature, pH, or exposure to chemicals, the protein structure may change, losing its shape without losing its primary sequence. This process is known as denaturation. Denaturation is often reversible because the primary structure of the polypeptide is conserved in the process if the denaturing agent is removed, allowing the protein to resume its function. Sometimes denaturation is irreversible, leading to loss of function. One example of irreversible protein denaturation is when an egg is fried. The albumin protein in the liquid egg white is denatured when placed in a hot pan when heat causes the strands of protein present to unravel and stick together in less ordered blobs. Not all proteins are denatured at high temperatures; for instance, bacteria that survive in hot springs have proteins that function at temperatures close to boiling. The stomach is also very acidic, has a low pH, and denatures proteins as part of the digestion process; however, the digestive enzymes of the stomach retain their activity under these conditions.
Protein folding is critical to its function. It was originally thought that the proteins themselves were responsible for the folding process. Only recently was it found that often they receive assistance in the folding process from protein helpers known as chaperones (or chaperonins) that associate with the target protein during the folding process. They act by preventing aggregation of polypeptides that make up the complete protein structure, and they disassociate from the protein once the target protein is folded.
Khan Academy link
CARBOHYDRATES
Most people are familiar with carbohydrates, especially when it comes to what we eat. Carbohydrates are an essential part of our diet; grains, fruits, and vegetables are all natural sources. More generally, carbohydrates are one of the four classes of macromolecules that compose all cells. While we may be most familiar with the role carbohydrates play in nutrition, they also have a variety of other essential functions in humans, animals, plants and bacteria. In this section we will discuss and review basic concepts of carbohydrate structure and nomenclature, as well as a variety of functions they play in cells.
Molecular Structures
In their simplest form, carbohydrates can be represented by the stoichiometric formula (CH2O)n, where n is the number of carbons in the molecule. For simple carbohydrates the ratio of carbon to hydrogen to oxygen in the molecule is 1:2:1. This formula also explains the origin of the term “carbohydrate”: the components are carbon (“carbo”) and the components of water (hence, “hydrate”). Simple carbohydrates are classified into three subtypes: monosaccharides, disaccharides, and polysaccharides, which will be discussed below. While simple carbohydrates fall nicely into this 1:2:1 ratio, carbohydrates can also be more complex, structurally. For example, many carbohydrates contain functional groups (remember them from our basic discussion about chemistry) besides the obvious hydroxyl. For example, carbohydrates can have phosphates, or amino groups substituted at a variety of sites within the molecule. These additional groups can provide additional properties to the molecule and will alter its overall function. However, even with these type of substitutions, the basic overall structure of the carbohydrate is retained and easily identified.
Nomenclature
One issue with carbohydrate chemistry is the nomenclature. Here are a few quick and simple rules:
- Simple carbohydrates end with an "...ose"; such as glucose, lactose, or dexrose
- Simple carbohydrates can be classified based on the number of carbon atoms in the molecule, such as a triose (3-carbons), pentose (5-carbons)or hexose (6-carbons).
- Simple carbohydrates can be classified based on the functional group found in the molecule, such as either a ketoses or aldoses.
- Polysaccharides are often organized by the number of sugar molecules in the chain, such as a monosaccharide, disaccharide or trisaccharide.
For a short video on carbohydrate classification see the Khan academy video (10 minutes in length) by clicking here.
Monosaccharides
Monosaccharides (mono- = “one”; sacchar- = “sweet”) are simple sugars, the most common of which is glucose. In monosaccharides, the number of carbons usually ranges from three to seven. If the sugar has an aldehyde group (the functional group with the structure R-CHO), it is known as an aldose, and if it has a ketone group (the functional group with the structure RC(=O)R'), it is known as a ketose.
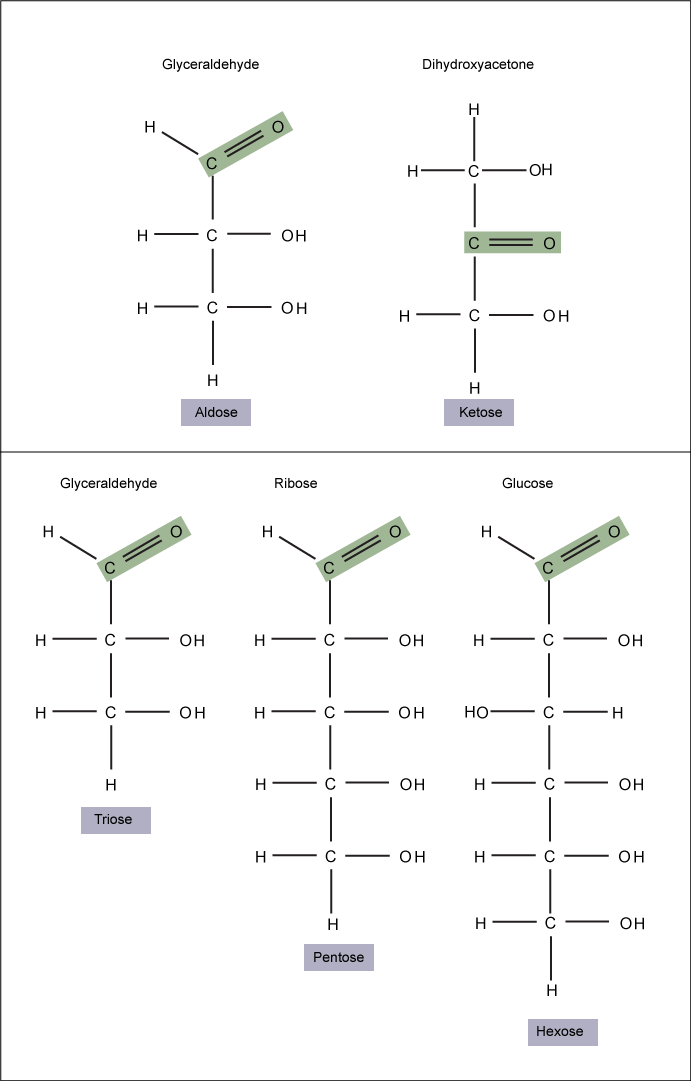
Monosaccharides are classified based on the position of their carbonyl group and the number of carbons in the backbone. Aldoses have a carbonyl group (indicated in green) at the end of the carbon chain, and ketoses have a carbonyl group in the middle of the carbon chain. Trioses, pentoses, and hexoses have three, five, and six carbon backbones, respectively.
Attribution: Marc T. Facciotti (own work)
Glucose versus Galactose
Galactose (part of lactose, or milk sugar) and glucose (found in sucrose, glucose disaccharride) are other common monosaccharides. The The chemical formula for glucose and galactose is C6H12O6, both are hexoses, but the arrangements of the hydrogens and hydroxyl groups are different at position C4. Because of this small difference, they differ structurally and chemically (and are known as chemical isomers) because of the different arrangement of functional groups around the asymmetric carbon; both of these monosaccharides have more than one asymmetric carbon (compare the structures in the Figure below).
Glucose, Galactose versus Fructose
A second comparison can be made when looking at glucose, galactose and fructose (the second carbohydrate that with glucose makes up the disaccharide sucrose, and is a common sugar found in fruit, hence the name). All three are hexoses, however, there is a major structural difference between glucose and glactose versus fructose; the carbon that contains the carbonyl (C=O). In glucose and galactose, it is on the C1 carbon forming and aldehyde group; in fructose it is on the C2 carbon, forming a keto group. The former are called aldoses based on the aldehyde group that is formed; while the later are designated ketoses based on the keto group. Again, this difference gives fructose different chemical and structural properties from the aldoses, glucose and galactose, though all three have the same chamical composition C6H12O6.
Glucose, galactose, and fructose are all hexoses. They are structural isomers, meaning they have the same chemical formula (C6H12O6) but a different arrangement of atoms.Glucose, galactose, and fructose
Linear versus Ring form of the charbohydrates
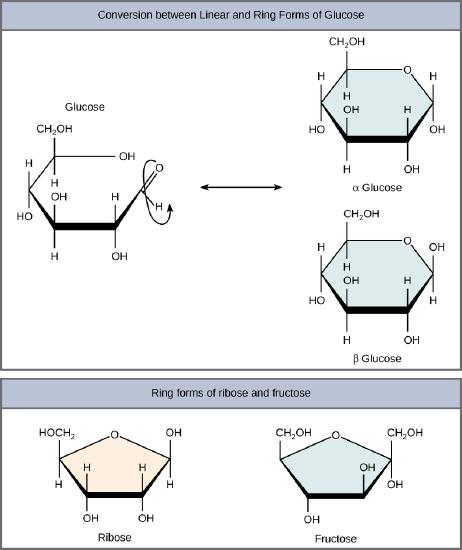
Monosaccharides can exist as a linear chain or as ring-shaped molecules; in aqueous solutions they are usually found in ring forms . Glucose in a ring form can have two different arrangements of the hydroxyl group (OH) around the anomeric carbon (carbon 1 that becomes asymmetric in the process of ring formation). If the hydroxyl group is below carbon number 1 in the sugar, it is said to be in the alpha (α) position, and if it is above the plane, it is said to be in the beta (β) position.
Five and six carbon monosaccharides exist in equilibrium between linear and ring forms. When the ring forms, the side chain it closes on is locked into an α or β position. Fructose and ribose also form rings, although they form five-membered rings as opposed to the six-membered ring of glucose.
Disaccharides
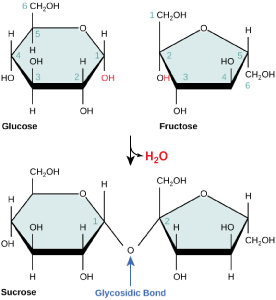
Disaccharides (di- = “two”) form when two monosaccharides undergo a dehydration reaction (also known as a condensation reaction or dehydration synthesis). During this process, the hydroxyl group of one monosaccharide combines with the hydrogen of another monosaccharide, releasing a molecule of water and forming a covalent bond. A covalent bond formed between a carbohydrate molecule and another molecule (in this case, between two monosaccharides) is known as a glycosidic bond. Glycosidic bonds (also called glycosidic linkages) can be of the alpha or the beta type.
Sucrose is formed when a monomer of glucose and a monomer of fructose are joined in a dehydration reaction to form a glycosidic bond. In the process, a water molecule is lost. By convention, the carbon atoms in a monosaccharide are numbered from the terminal carbon closest to the carbonyl group. In sucrose, a glycosidic linkage is formed between carbon 1 in glucose and carbon 2 in fructose.
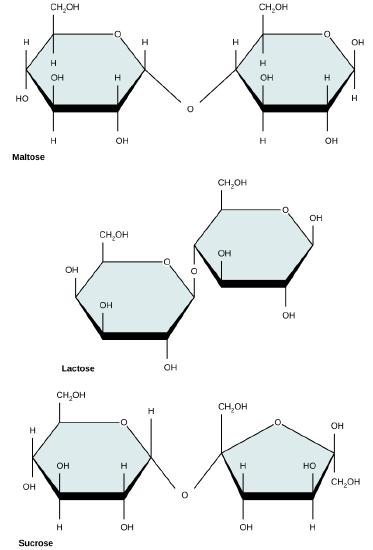
Common disaccharides include lactose, maltose, and sucrose. Lactose is a disaccharide consisting of the monomers glucose and galactose. It is found naturally in milk. Maltose, or malt sugar, is a disaccharide formed by a dehydration reaction between two glucose molecules. The most common disaccharide is sucrose, or table sugar, which is composed of the monomers glucose and fructose.
Common disaccharides include maltose (grain sugar), lactose (milk sugar), and sucrose (table sugar).
Interactions with carbohydrates
We have just discussed the various types and structures of carbohydrates found in biology. The next thing to address is how these compounds interact with other compounds. The answer to that is that it depends on the final structure of the carbohydrate. Because carbohydrates have many hydroxyl groups associated with the molecule, they are therefore excellent H-bond donors and acceptors . Monosaccharides can quickly and easily form H-bonds with water and are readily soluble. All of those H-bonds also make them quite "sticky". This also true for many disaccharides and many short chain polymers. Longer polymers, may not be so readily soluble.
Finally, the ability to form a variety of H-bonds, allows polymers of carbohydrates or polysaccharides to form strong intra-molecular and inter-molocular bonds. In a polymer, because there are so many H-bonds, this can provide alot of strength to the molecule or molecular complex, especially if the polymers interact. Just think of cellulose, a polymer of glucose, if you have any doubts.
Polysaccharides
A long chain of monosaccharides linked by glycosidic bonds is known as a polysaccharide (poly- = “many”). The chain may be branched or unbranched, and it may contain different types of monosaccharides. The molecular weight may be 100,000 daltons or more depending on the number of monomers joined. Starch, glycogen, cellulose, and chitin are primary examples of polysaccharides.
Starch is the stored form of sugars in plants and is made up of a mixture of amylose and amylopectin (both polymers of glucose). Plants are able to synthesize glucose, and the excess glucose, beyond the plant’s immediate energy needs, is stored as starch in different plant parts, including roots and seeds. The starch in the seeds provides food for the embryo as it germinates and can also act as a source of food for humans and animals. The starch that is consumed by humans is broken down by enzymes, such as salivary amylases, into smaller molecules, such as maltose and glucose. The cells can then absorb the glucose.
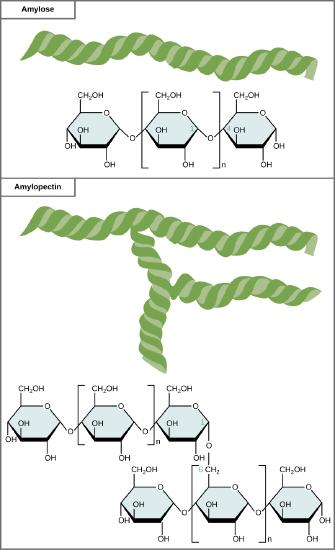
Starch is made up of glucose monomers that are joined by α 1-4 or α 1-6 glycosidic bonds. The numbers 1-4 and 1-6 refer to the carbon number of the two residues that have joined to form the bond. As illustrated below, amylose is starch formed by unbranched chains of glucose monomers (only α 1-4 linkages), whereas amylopectin is a branched polysaccharide (α 1-6 linkages at the branch points).
Amylose and amylopectin are two different forms of starch. Amylose is composed of unbranched chains of glucose monomers connected by α 1,4 glycosidic linkages. Amylopectin is composed of branched chains of glucose monomers connected by α 1,4 and α 1,6 glycosidic linkages. Because of the way the subunits are joined, the glucose chains have a helical structure. Glycogen (not shown) is similar in structure to amylopectin but more highly branched.
Glycogen is the storage form of glucose in humans and other vertebrates and is made up of monomers of glucose. Glycogen is the animal equivalent of starch and is a highly branched molecule usually stored in liver and muscle cells. Whenever blood glucose levels decrease, glycogen is broken down to release glucose in a process known as glycogenolysis.
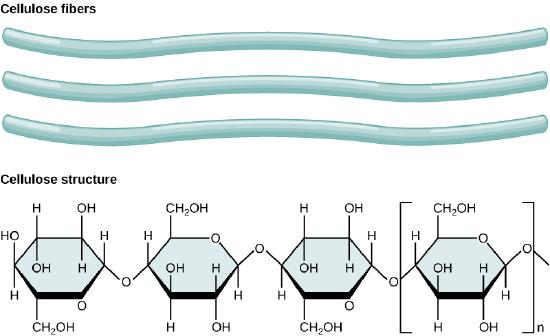
Cellulose is the most abundant natural biopolymer. The cell wall of plants is mostly made of cellulose; this provides structural support to the cell. Wood and paper are mostly cellulosic in nature. Cellulose is made up of glucose monomers that are linked by β 1-4 glycosidic bonds.
In cellulose, glucose monomers are linked in unbranched chains by β 1-4 glycosidic linkages. Because of the way the glucose subunits are joined, every glucose monomer is flipped relative to the next one resulting in a linear, fibrous structure.
Note:
Possible discussion: Cellulose is not very soluble in water when in a crystalline state - that can be approximated by the stacked cellulose fiber depiction above in the figure above. Can you suggest a reason for why (types of interactions) it might be so insoluble?
As shown in the figure above, every other glucose monomer in cellulose is flipped over, and the monomers are packed tightly as extended long chains. This gives cellulose its rigidity and high tensile strength—which is so important to plant cells. While the β 1-4 linkage cannot be broken down by human digestive enzymes, herbivores such as cows, koalas, buffalos, and horses are able, with the help of the specialized flora in their stomach, to digest plant material that is rich in cellulose and use it as a food source. In these animals, certain species of bacteria and protists reside in the rumen (part of the digestive system of herbivores) and secrete the enzyme cellulase. The appendix of grazing animals also contains bacteria that digest cellulose, giving it an important role in the digestive systems of ruminants. Cellulases can break down cellulose into glucose monomers that can be used as an energy source by the animal. Termites are also able to break down cellulose because of the presence of other organisms in their bodies that secrete cellulases.
NUCLEIC ACIDS
Nucleic acids are molecules made up of nucleotides that carry the genetic blueprint of a cell. Each nucleotide is made up of a pentose sugar, a nitrogenous base, and a phosphate group. There are two types of nucleic acids: DNA and RNA. DNA carries the genetic blueprint of the cell and is passed on from parents to offspring. Double stranded DNA has a helical structure with the two strands running in opposite directions. The two strands are connected by hydrogen bonds, and chemically complementary to each other. Interactions known as "base stacking" interactions also help stabilize the double helix. RNA can either be single-stranded, or double-stranded and is made of a pentose sugar (ribose), a nitrogenous base, and a phosphate group. RNA is involved in protein synthesis as a messenger, the regulation of protein synthesis, other regulatory processes and some catalytic activity. Messenger RNA (mRNA) is copied from the DNA, is exported from the nucleus to the cytoplasm, and contains information for the construction of proteins. Ribosomal RNA (rRNA) is a part of the ribosomes at the site of protein synthesis, whereas transfer RNA (tRNA) carries the amino acid to the site of protein synthesis. microRNA regulates the use of mRNA for protein synthesis.
Nucleotide Structure
The two main types of nucleic acids are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). The main common difference between these two types of nucleic acids is the presence or absence of a hydroxyl group at the C2 position, also called the 2' position, of the ribose. DNA lacks the ribose and contains a hydrogen atom at that position, hence the name, "deoxy" ribonucleic acid whereas RNA has a hydroxyl functional group at that position.
DNA and RNA are made up of monomers known as nucleotides. Individual nucleotides condense with one another other to form a nucleic acid polymer. Each nucleotide is made up of three components: a nitrogenous base (for which there are five different types), a pentose (five-carbon) sugar, and a phosphate group. These are depicted below.
The Nitrogenous Base
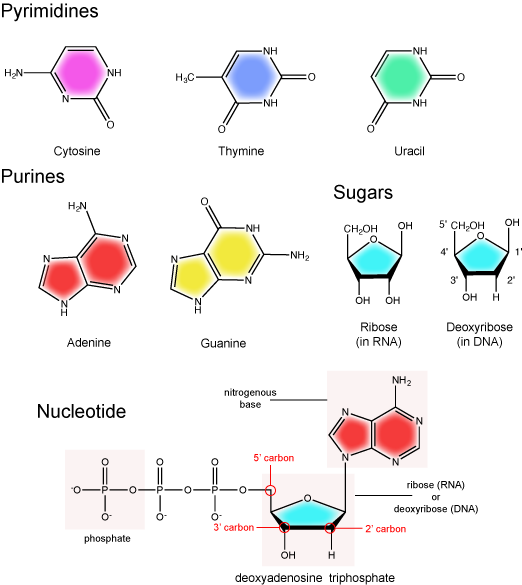
The nitrogenous bases of nucleotides are organic molecules and are so named because they contain carbon and nitrogen. They are bases because they contain an amino group that has the potential of binding an extra hydrogen, and thus acting as a base by decreases the hydrogen ion concentration in the local environment. Each nucleotide in DNA contains one of four possible nitrogenous bases: adenine (A), guanine (G) cytosine (C), and thymine (T). RNA contains adenine (A), guanine (G) cytosine (C), and uracil (U) instead of thymine (T).
Adenine and guanine are classified as purines. The primary distinguishing feature of the structure of a purine is double carbon-nitrogen ring. Cytosine, thymine, and uracil are classified as pyrimidines. These are distinguished structurally by a single carbon-nitrogen ring. You will be expected to recognize that each of these ring structures is decorated by functional groups that may be involved in a variety of chemistries and interactions.
Note: Practice
Take a moment to review the figure above of the nitrogenous base: Identify functional groups as described in class. For each functional group identified, describe what type of chemistry you expect it to be involved in. If hydrogen bonding, does the functional group act as a donor or acceptor?
The Pentose Sugar
The pentose sugar contains 5 carbon atoms. Each carbon atom of the sugar molecule are numbered as 1′, 2′, 3′, 4′, and 5′ (1′ is read as “one prime”). The two main functional groups that are attached to the sugar are often refured to in reference to the carbon number they are bound to. For example, the phosphate residue is attached to the 5′ carbon of the sugar and the hydroxyl group is attached to the 3′ carbon of the sugar. We will often use the carbon number to refer to functional groups on nucleotides so be very familiar with the structure of the pentose sugar.
The pentose sugar in DNA is called deoxyribose, and in RNA, the sugar is ribose. The difference between the sugars is the presence of the hydroxyl group on the 2' carbon of the ribose and its absence on the 2' carbon of the deoxyribose. Hence you can determine if you are looking at a DNA or RNA nucleotide by the presence or absence of the hydroxyl group on the 2' carbon atom - you will likely be asked to do so on numerous occasions (including exams).
The Phosphate Group
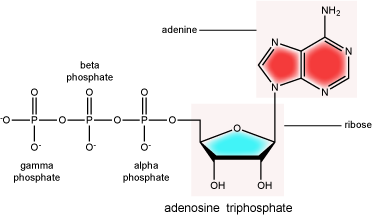
There can be anywhere between 1 and 3 phosphate groups bound to the 5' carbon of the sugar. When one phosphate bound the nucleotide is referred to as a Nucleotide MonoPhosphate (NMP). If 2 phosphates are bound the nucleotide is referred to as Nucleotide DiPhosphate (NDP). When 3 phosphates are bound to the nucleotide it is referred to as a Nucleotide TriPhosphate (NTP). The phosphoanhydride bonds between that link the phosphate groups to each other have specific chemical properties that make them good for various biological functions. The hydrolysis of the bonds between the phosphate groups is thermodynamically exergonic in biological conditions and nature has evolved numerous mechanisms to couple this negative change in free-energy to help drive many reactions in the cell. The figure below shows an example of the hydrolysis of the nucleotide triphosphate ATP.
Note: "High energy" bonds
The term "high energy bond" is used A LOT in biology. It is, however, one of those shortcuts we referred to earlier. The term refers to the amount of negative free energy associated with the HYDROLYSIS of that bond! The water is important. While we have tried to minimize the use of the vernacular "high energy" when referring to bonds, keep the above in mind when you are reading or listening to discussions in biology.
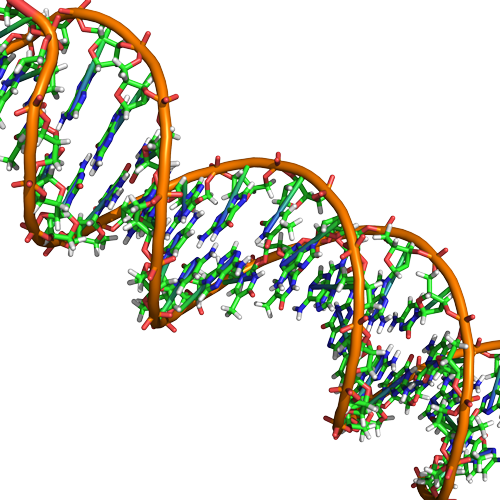
Double Helix Structure of DNA
DNA has a double-helix structure (shown below). The sugar and phosphate lie on the outside of the helix, forming the backbone of the DNA. The nitrogenous bases are stacked in the interior, like the steps of a staircase, in pairs; the pairs are bound to each other by hydrogen bonds. Every base pair in the double helix is separated from the next base pair by 0.34 nm. The two strands of the helix run in opposite directions, meaning that the 5′ carbon end of one strand will face the 3′ carbon end of its matching strand. This is referred to as antiparallel orientation.
In a double helix, certain combinations of base pairing are chemically more favored than others based on the types and locations of functional groups on the nitrogenous bases of each nucleotide. In biology we find that adenine (A) is chemically complementary with thymidine (T) and guanine (G) is chemically complementary with cytosine (C), as shown below. We often refer to this pattern as "base complementarity" and say that the antiparallel strands are complementary to each other. For example, if the sequence of one strand is of DNA is 5'-AATTGGCC-3', the complementary strand would have the sequence 5'-GGCCAATT-3'.

Functions and roles of nucleic acids and nucleotides
Nucleic acids play a variety of roles in in cellular process besides being the information storage molecule. Nucleic acids, RNA in particular, are believed to be the first biologically active molecules during a period referred to as the "RNA world" when catalytic RNA were thought to serve the dual role as catalysts and information storing molecules. Remnants of the RNA world can be seen in many riboprotein complexes essential for life. In these RNA-Protein complexes the RNA serves both catalytic and structural roles. Examples include of such complexes include, ribosomes, RNases, splicesosome complexes and Telomerase. Nucleotides such as ATP and GTP also serve as mobile short-term energy transport units for the cell. Nucleotides also play important roles as co-factors (in addition to energy vehicles) for many enzymatic reactions. Like lipids, proteins and carbohydrates, nucleic acids and nucleotides play a wide variety of roles in the cell.
Lipids
Lipids include a diverse group of compounds. Many lipids are at their core hydrocarbons, molecules that include many nonpolar carbon–carbon or carbon–hydrogen bonds. The abundance of non polar functional groups give lipids a degree of hydrophobic (“water fearing”) character and most lipids have low solubilities in water. Lipids include molecules like fats, oils, waxes, phospholipids, and steroids. They perform many different important functions in biology (energy storage, insulation, act as barrier, signaling). The diversity of lipid molecules and their range of biological activities is also large - perhaps surprisingly so to an new student of biology. Let's explore a bit.
Fats and Oils
A common fat molecule or triglyceride is a molecule derived from two types of molecular components — a polar "head" group and a non-polar "tail" group. Glycerol, a carbohydrate, is an organic compound composed of three carbons, five hydrogens, and three hydroxyl (OH) groups. The "head" group of a triglyceride is derived from a single glycerol molecule. The non-polar fatty acid "tail" group consists of three hydrocarbons (a functional group composed of C-H bonds) that also have a polar carboxyl functional group (hence the term "fatty acid" - the carboxyl group is acidic at most biologically relevant pHs). The number of carbons in the fatty acid may range from 4 to 36; most common are those containing 12–18 carbons. These types of molecules are generally hydrophobic and while they have numerous functions are probably best known for their roles in body fat and plant oils.
Note: Possible Discussion
The models of the triglycerides shown above show the relative positions of the atoms in the molecule. If you Google for images of triglycerides you will find some models that show the phospholipid tails in wildly different positions. What is equally well represented by the model in the book and others on-line? Using your intuition, give an opinion for which model you think is a more correct representation of real-life. Why?
Stearic acid is a common saturated fatty acid, oleic acid and linolenic acid are common unsaturated fatty acids.
Attribution: Marc T. Facciotti (own work)
Note: Possible Discussion
The physical properties of different natural fats like butter, canola oil, etc. (composed mostly of triglycerides) are very dependent on two factors:
1) The number of carbons in the hydrocarbon chains
2) The number of desaturations (double bonds) in the carbon chains.
The first factor influences how these molecules interact with each other and with water while the second factor influences their shape dramatically. The introduction of a double bond causes a "kink" in the otherwise relatively "straight" hydrocarbon (depicted in a slightly exaggerated figure below). Based on what you can surmise from this brief description, propose a rationale - in your own words - to explain why butter is solid at room temperature while vegetable oil is liquid.
Fact: butter has a greater percentage of longer and saturated hydrocarbons in its triglycerides than does vegetable oil.
Sterols
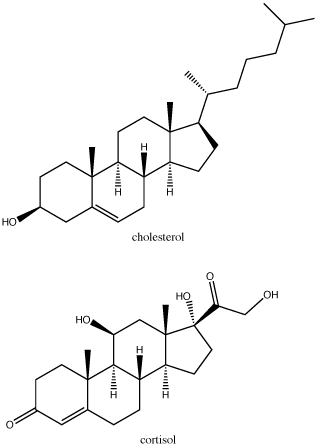
Steroids are lipids with a fused ring structure. Although they do not resemble the other lipids discussed here, they are designated a lipid because they are also largely composed of carbons and hydrogens, are hydrophobic and insoluble in water. All steroids have four linked carbon rings and several of them, like cholesterol, have a short tail. Many steroids also have the –OH functional group, which puts them in the alcohol classification (sterols). Cholesterol is the most common steroid. Cholesterol is mainly synthesized in the liver and is the precursor to many steroid hormones such as testosterone and estradiol, which are secreted by the gonads and endocrine glands. It is also the precursor to Vitamin D. Cholesterol is also the precursor of bile salts, which help in the emulsification of fats and their subsequent absorption by cells. Although cholesterol is often spoken of in negative terms, it is necessary for proper functioning of many animal cells. It is a component of the plasma membrane of animal cells and is thought to play a variety of roles that stem from its influence on modulating membrane structure, organization and fluidity.
Note: Possible Discussion
In the molecule of cortisol above: what parts of the molecule would you classify as functional groups? Is there any disagreement over what should and should not be included as a functional group?
Phospholipids
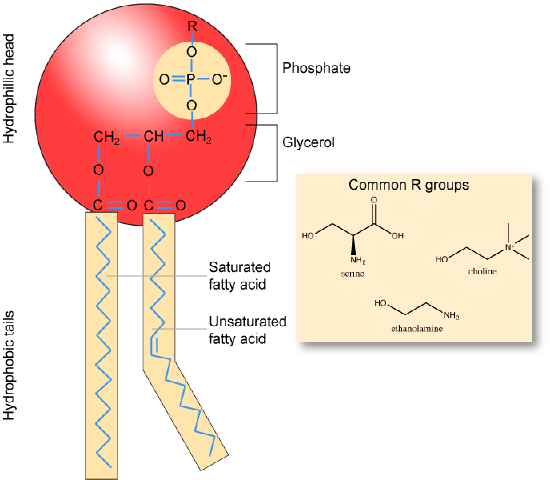
Phospholipids are major constituents of the cell membrane, the outermost layer of cells. Like fats, they are composed of fatty acid chains attached to a polar head group. Specifically, there are two fatty acid tails and a phosphate group as the polar head group. The phospholipid is an amphipathic molecule, meaning it has a hydrophobic part and a hydrophilic part. The fatty acid chains are hydrophobic and cannot interact with water, whereas the phosphate-containing head group is hydrophilic and interacts with water. Can you identify the functional groups on the phospholipid below that give each part of the phospholipid its properties?
Note
Make sure to note in the figure below that the phosphate group has an R-group linked to one of the oxygen atoms. R is a variable commonly used in these types of diagrams to indicate that some other atom or molecule is bound at that position. That part of the molecule can be different in different phospholipids - and will impart some different chemistry to the whole molecule. At the moment, however, you are responsible for being able to recognize this type of molecule (no matter what the R group is) because of the common core elements - the glycerol backbone, the phosphate group, and the two hydrocarbon tails.
A phospholipid bilayer forms as the basic structure of the cell membrane. The fatty acid tails of phospholipids face inside, away from water, whereas the phosphate group faces outside, hydrogen bonding with water. Phospholipids are responsible for the dynamic nature of the plasma membrane. The phospholipids will spontaneously form a structure known as a micelle, where the hydrophilic phosphate heads face the outside and the fatty acids face the interior of this structure.
Note: Possible Discussion
Above it says that if you were to take some pure phospholipids and drop them into water that some if it would spontaneously (on its own) form into micelles. This sounds a lot like something that could be described by an energy story. Go back to the energy story rubric and try to start creating an energy story for this process - I expect that the steps involving the description of energy might be difficult at this point (we'll come back to that later) but you should be able to do at least the first three steps. You can constructively critique (politely) each other's work to create an optimized story.
The phospholipid membrane is discussed in detail in a later module. It will be important to remember the chemical properties associated with the functional groups in the phospholipid in order to understand the function of the cell membrane.
For additional information on lipids:
For an additional perspective on lipids, explore the interactive animation “Biomolecules: The Lipids”. For more information on lipids, please visit the UCD Chemwiki site at Chemwiki lipids
Another perspective on lipids, that contains a variety of animations to help you, is the following link from Carnegie Mellon University, Department of Biological Sciences flash tutorial on lipids.