5.1: Introduction
- Page ID
- 15978
OBjectives
- Learn how to use differential and selective media to identify bacterial species.
- Be able to perform the following biochemical tests and understand how they work: carbohydrate fermentation, gelatin hydrolysis, starch hydrolysis, casein hydrolysis, urea hydrolysis, citrate utilization, catalase activity, tryptophan hydrolysis (indole test).
- Be able to relate the results of the tests performed to the different types of metabolic activities carried out by individual bacterial species.
As you observed in Labs 3 and 4, staining procedures can be used to discriminate between bacteria based on morphology, arrangement, cell wall structure, and their possession of other structures associated with bacterial cells such as flagella, capsules and spores. However, staining procedures are not sufficient to identify bacteria at the species level. For example, E. coli, P. aeruginosa, P. vulgaris, E. aerogenes, and many other bacteria are all Gram negative rods and are indistinguishable by Gram staining. Therefore, additional methods are required to identify individual species.
Even though different bacterial species may appear identical under the microscope, they all have a different genetic makeup (different DNA), and therefore produce different types of enzymes that allow them to carry out a characteristic set of biochemical reactions. In other words, each bacterial species has a characteristic metabolism that can be used to distinguish them from other species, and therefore unknown bacteria are often identified based on both their appearance under the microscope as well as their metabolic properties.
Examples of differences in metabolic properties were observed in Lab 2, when you inoculated E. coli and P. aeruginosa onto EMB agar. Each of these bacteria produced different colored colonies on this media, depending on their ability to ferment lactose. In addition to EMB and MS, there are many other types of differential media that can be used in the identification of bacteria.
When using differential media, it is always important to remember that reactions in these media can vary based on the incubation time, the incubation temperature, the number of bacteria in the initial inoculum and how the medium is inoculated, among other variables. For example, an inoculum containing large amounts of bacteria might give a strong positive reaction, while one containing a very small number of bacteria might only produce a weakly positive, or perhaps a negative reaction. It is also important to remember that not all bacterial species will give definitive positive or negative results on these media. Since not all differential media will provide meaningful results for all bacterial species, it is important to choose the media carefully when identifying unknowns.
In many metabolic tests, end products are produced that change the pH of the medium. To measure this pH change, pH indicators (chemicals that change color depending on pH) are included in the medium. Some common pH indicators are phenol red, bromocresol purple, and bromothymol blue. Each pH indicator has a range of pH values over which it changes color (see below).
| pH Indicator | |||
| phenol red | < pH 6.8 = yellow | pH 6.8 – 7.4 = red | pH >7.4 = pink/magenta |
| bromocresol purple | < pH 6.8 = yellow | > 6.8 pH = purple | |
| bromothymol blue | < pH 6.0 = yellow | pH 6.1 – 7.5 = green | pH >7.5 = blue |
Note
Color pictures showing the results for all metabolic tests can be found on the Microbiology Review site on Blackboard.
A. Carbohydrate Fermentation
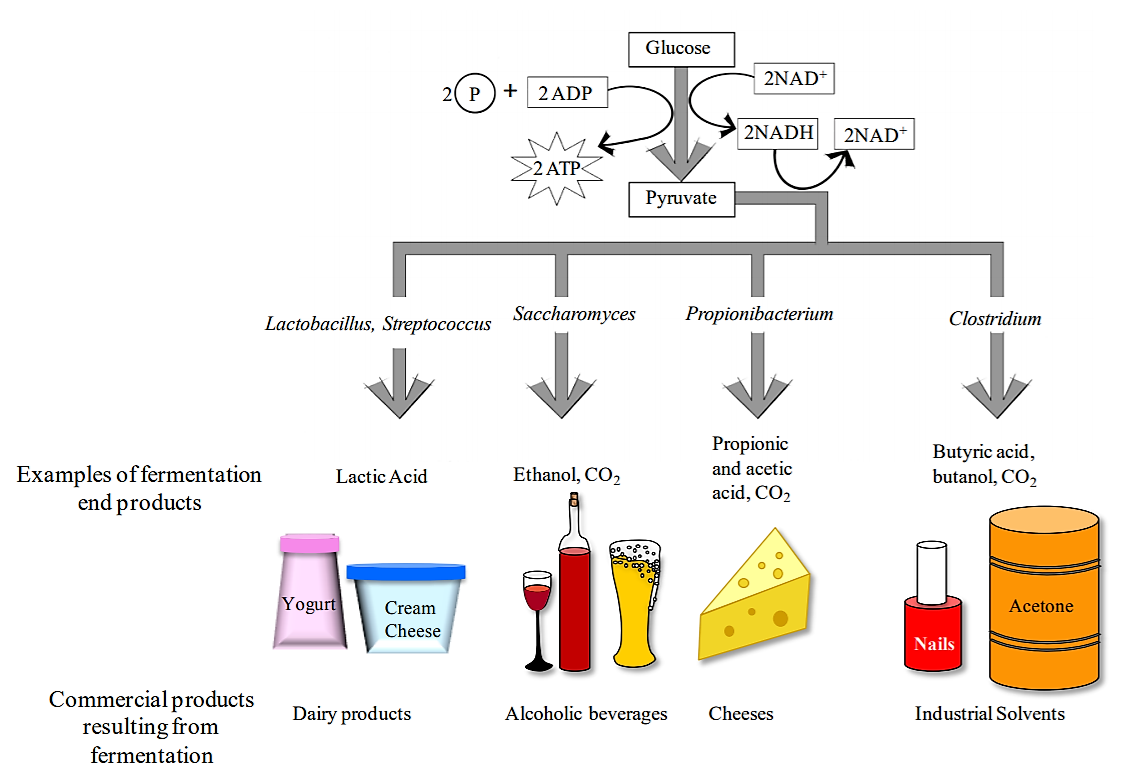
Fermentation is a metabolic process that some bacteria use to break down glucose when O2 is not available. Fermentation includes the reactions of glycolysis (where a single molecule of glucose is broken down into 2 molecules of pyruvate), as well as additional reactions that produce a variety of end products (acids, alcohols, gases). The end products are characteristic of individual bacterial species.
.png?revision=1&size=bestfit&width=859&height=591)
Although the ultimate substrate molecule for fermentation is always glucose, some bacteria use additional chemical reactions to convert other monosaccharides as well as disaccharides into glucose. Therefore bacteria can be differentiated both based on their ability to ferment various carbohydrates, as well as the end products that result from the fermentation process.
The medium used to test carbohydrate fermentation is a nutrient broth that contains a fermentable carbohydrate (usually a monosaccharide or a disaccharide), peptone (amino acids) as well as a pH indicator. The pH of the medium is adjusted to approximately 7.5, so it appears orange/red when using phenol red. If the carbohydrate in the medium is fermented and acidic end products are formed, a color change to yellow will result. Occasionally, bacteria will not ferment the carbohydrate, but instead will break down proteins producing ammonia (NH3) in the growth medium. In this case, the medium will become more alkaline.
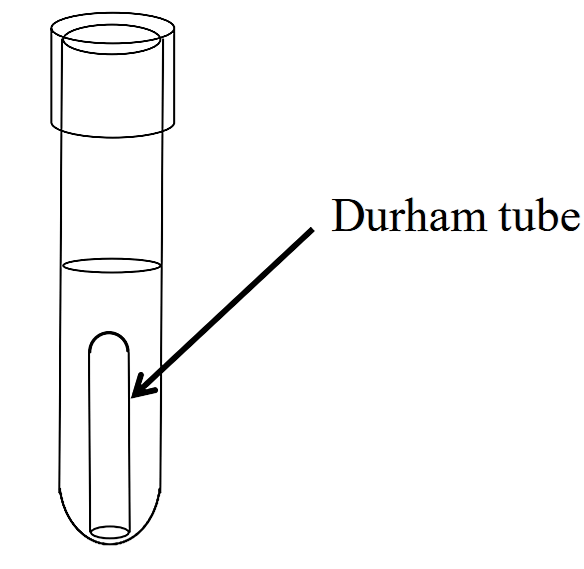
Some bacteria will produce gases when fermenting a carbohydrate. To detect these gases, a Durham tube is used. This is a small inverted tube that is placed within the larger glass tube containing the fermentation medium (see below). If gases (typically CO2) are produced during the fermentation process, a bubble will form at the top of the Durham tube. If you see a bubble in the Durham tube, the medium will also be acidic.
.png?revision=1&size=bestfit&width=439&height=428)
Carbohydrate fermentation media are often used to differentiate members of the family Enterobacteriaceae (e.g., Escherichia coli, Enterobacter aerogenes) from each other.
B. Gelatin Hydrolysis
Gelatin is a protein derived from the collagen found in connective tissue of bones, skin and tendons. Robert Koch first used gelatin as a solidifying agent for growth media, but switched to agar when it was realized that gelatin does not stay solid at temperatures above 28oC and that some bacteria have the ability to chemically break it down. Some species of bacteria secrete an extracellular enzyme (exoenzyme) called gelatinase (a type of proteinase), that breaks gelatin down into its constituent polypeptides and amino acids. The small amino acids can then be brought into the bacterial cell. This is why agar is now used as the solidification agent in bacterial growth media. Since gelatinase breaks down connective tissue, it is sometimes called “spreading factor” and contributes to the virulence of a pathogen.
.png?revision=1&size=bestfit&width=484&height=118)
The presence of gelatinase can be detected by using nutrient gelatin deeps. This medium contains gelatin and nutrients for bacterial growth. A positive result is indicated by the gelatin becoming liquefied due to the enzyme activity. It is important to note that if the nutrient gelatin deeps are incubated at 37°C, then the medium must be cooled to below 25°C before a result can be recorded. A negative control is therefore very important in this experiment.
C. Starch Hydrolysis
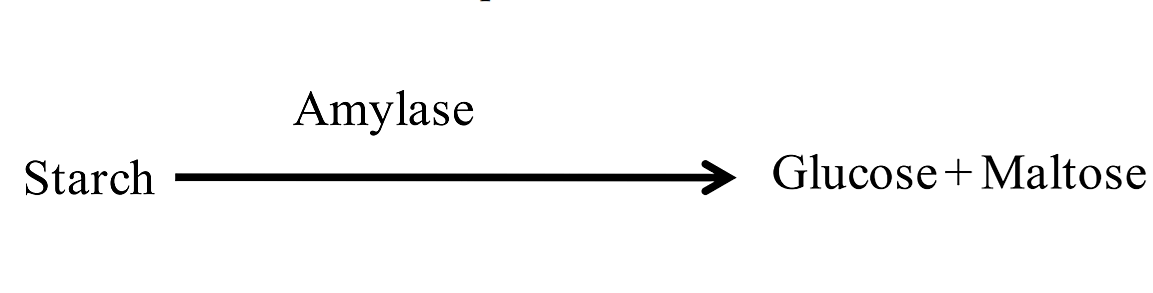
Starch is a polymer of glucose that is too large to be transported into bacterial cells. For bacteria to be able to use the glucose in starch as an energy source, the macromolecule must be broken down extracellularly into smaller monosaccharide and disaccharide subunits. Some bacteria secrete the extracellular enzyme (exoenzyme) amylase that breaks down starch into monosaccharides and disaccharides (glucose and maltose). These smaller molecules can then be transported inside the bacterial cell.
.png?revision=1&size=bestfit&width=339&height=87)
Starch agar is used to determine if bacteria can produce the amylase enzyme. Bacteria that secrete amylase will break down the starch in the medium surrounding the bacterial colony. After the plate is inoculated and incubated, iodine is poured on the plate. The combination of iodine and starch results in the formation of a dark blue-black color. Regions of the agar where no starch is present (i.e., where starch has been broken down by amylase) will remain the color of the iodine (light yellow or gold).
D. Casein Hydrolysis
Some bacteria secrete enzymes called proteinases that break down proteins. You have already read about one kind of proteinase—gelatinase. Milk contains large proteins called caseins. Some bacteria secrete caseinases that break down casein outside of the bacterial cell so the smaller products (e.g., amino acids) can be transported inside the cell and further metabolized.
Milk agar (which contains powdered milk) is used to detect the presence of bacterial caseinases. This medium is cloudy because when milk is mixed with agar, the casein forms a colloid through which light cannot pass. The presence of caseinases can be detected by observing a clearing in the agar around the bacterial growth, which indicates that the caseins have been broken down into transparent end products (amino acids and peptides), which are then taken up by the cells.
E. Urea Hydrolysis
Urea is a common waste product that results from the breakdown of proteins. Urea agar slants are used to test for the presence of a urease enzyme that is produced by some bacteria. Bacteria that produce ureases can break urea down into ammonia and carbon dioxide (see below). The ammonia that results from the breakdown of urea causes the medium of the slant to become more alkaline; this pH change is detected by the phenol red, which changes to a hot pink color. Organisms that do not break down urea may grow on the slant, but the slant will remain its original color, or will become more yellow (due to the production of acids).
.png?revision=1&size=bestfit&width=339&height=132)
F. Citrate Utilization
Organisms that can survive using citrate as the sole source of carbon have a citrate permease enzyme that can transport citrate molecules into the cell. The citrate is then made into pyruvate, which can be converted into a variety of different products. Simmon’s citrate is a chemically defined medium that contains sodium citrate as the sole carbon source as well as the pH indicator bromothymol blue. Bacteria that can grow on this medium (i.e., that can survive on citrate as the sole source of carbon) produce alkaline byproducts that will change the medium from green (neutral pH) to blue (alkaline pH).
G. Catalase Activity
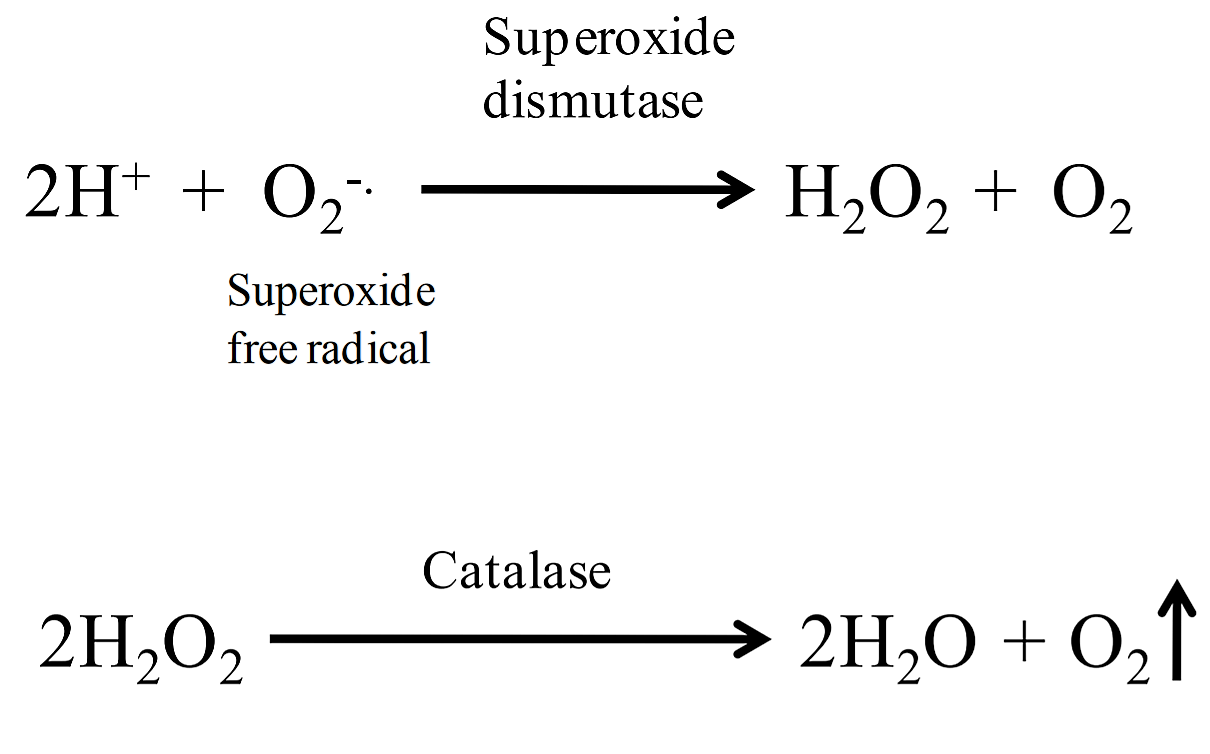
Byproducts of aerobic metabolism include two toxic compounds: superoxide free radicals (2O2 -) and hydrogen peroxide (H2O2). To remove these compounds, cells produce enzymes to break them down. Cells can convert superoxide free radicals to hydrogen peroxide by using the enzyme superoxide dismutase (SOD); catalase breaks down hydrogen peroxide into water and oxygen.
.png?revision=1&size=bestfit&width=345&height=215)
A simple test to determine if bacteria produce catalase is to add hydrogen peroxide to bacteria on an agar slant or to bacteria spread on a slide. If catalase is present, the hydrogen peroxide will be broken down into water and oxygen gas, resulting in the production of bubbles. This test does not require any special type of medium, however it should never be performed on organisms that have been grown on blood agar (a medium that contains blood). This is because there is a catalase activity in blood that would produce a false positive result. Most aerobic and facultatively anaerobic organisms produce SOD and catalase (note: some species use peroxidase rather than catalase to break down hydrogen peroxide). Obligate anaerobes lack these enzymes, which is why they cannot survive in an atmosphere containing oxygen.
H. Tryptophan Hydrolysis:
The Indole Test Tryptophan is an amino acid found in most proteins. Some bacteria produce tryptophanase, an enzyme that breaks tryptophan down into indole, ammonia and pyruvate (see below). The pyruvate and ammonia are converted into other molecules, but the indole accumulates, and thus can be detected in the media.
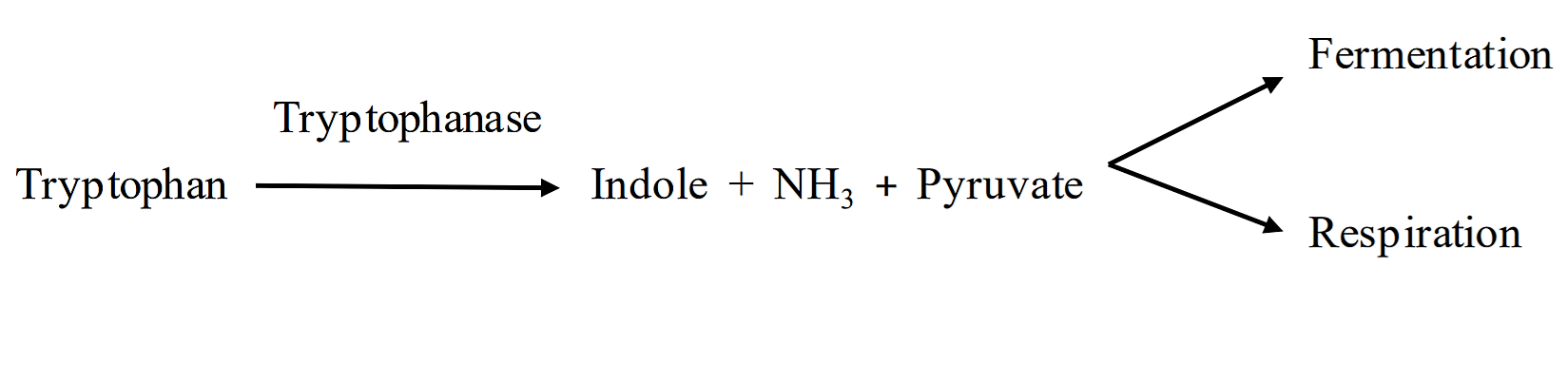.png?revision=1&size=bestfit&width=463&height=113)
The presence of indole indicates that an organism produces the enzyme tryptophanase. Indole can be detected using a chemical known as Kovac’s reagent. We will be using a BBL dry slide to detect indole. This is a disposable slide that employs a dry reagent slide impregnated with Kovac’s reagent. The slide is divided into four large segments to enable multiple tests on one slide. The presence of indole will cause a pink color to develop on the dry-slide; no color change will occur if no indole is detected.
Note
For the indole test to be performed, the organism must be grown using medium containing tryptophan.
Indole + Kovac’s reagent + HCl + Amyl Alcohol →Rosindole dye (pink-red)
Key Terms
fermentation, amylase, gelatinase, caseinase, urease, citrate permease, catalase, indole, tryptophanase, Kovac’s reagent, BBL Dry Slide, agar slant (locate butt and slant), Durham tube, Simmon’s citrate


