W2017_Lecture_18_reading
- Page ID
- 21362
NUCLEIC ACIDS
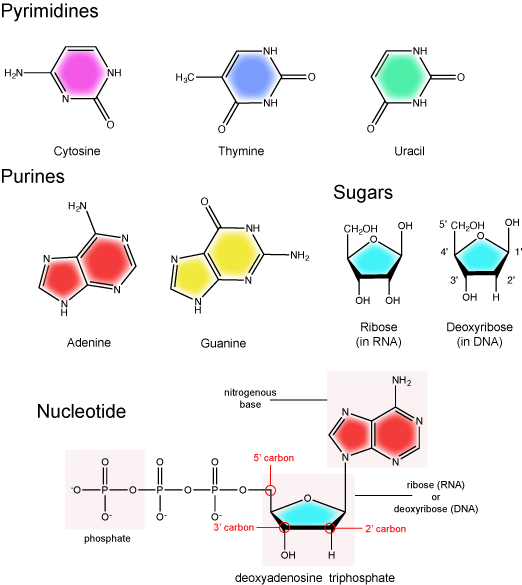
Nucleic acids are molecules made up of nucleotides that carry the genetic blueprint of a cell. Each nucleotide is made up of a pentose sugar, a nitrogenous base, and a phosphate group. There are two types of nucleic acids: DNA and RNA. DNA carries the genetic blueprint of the cell and is passed on from parents to offspring. Double stranded DNA has a helical structure with the two strands running in opposite directions. The two strands are connected by hydrogen bonds, and chemically complementary to each other. Interactions known as "base stacking" interactions also help stabilize the double helix. RNA can either be single-stranded, or double-stranded and is made of a pentose sugar (ribose), a nitrogenous base, and a phosphate group. RNA is involved in protein synthesis as a messenger, the regulation of protein synthesis, other regulatory processes and some catalytic activity. Messenger RNA (mRNA) is copied from the DNA, is exported from the nucleus to the cytoplasm, and contains information for the construction of proteins. Ribosomal RNA (rRNA) is a part of the ribosomes at the site of protein synthesis, whereas transfer RNA (tRNA) carries the amino acid to the site of protein synthesis. microRNA regulates the use of mRNA for protein synthesis.
Nucleotide Structure
The two main types of nucleic acids are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). The main common difference between these two types of nucleic acids is the presence or absence of a hydroxyl group at the C2 position, also called the 2' position, of the ribose. DNA lacks the ribose and contains a hydrogen atom at that position, hence the name, "deoxy" ribonucleic acid whereas RNA has a hydroxyl functional group at that position.
DNA and RNA are made up of monomers known as nucleotides. Individual nucleotides condense with one another other to form a nucleic acid polymer. Each nucleotide is made up of three components: a nitrogenous base (for which there are five different types), a pentose (five-carbon) sugar, and a phosphate group. These are depicted below.
The Nitrogenous Base
The nitrogenous bases of nucleotides are organic molecules and are so named because they contain carbon and nitrogen. They are bases because they contain an amino group that has the potential of binding an extra hydrogen, and thus acting as a base by decreases the hydrogen ion concentration in the local environment. Each nucleotide in DNA contains one of four possible nitrogenous bases: adenine (A), guanine (G) cytosine (C), and thymine (T). RNA contains adenine (A), guanine (G) cytosine (C), and uracil (U) instead of thymine (T).
Adenine and guanine are classified as purines. The primary distinguishing feature of the structure of a purine is double carbon-nitrogen ring. Cytosine, thymine, and uracil are classified as pyrimidines. These are distinguished structurally by a single carbon-nitrogen ring. You will be expected to recognize that each of these ring structures is decorated by functional groups that may be involved in a variety of chemistries and interactions.
Note: Practice
Take a moment to review the figure above of the nitrogenous base: Identify functional groups as described in class. For each functional group identified, describe what type of chemistry you expect it to be involved in. If hydrogen bonding, does the functional group act as a donor or acceptor?
The Pentose Sugar
The pentose sugar contains 5 carbon atoms. Each carbon atom of the sugar molecule are numbered as 1′, 2′, 3′, 4′, and 5′ (1′ is read as “one prime”). The two main functional groups that are attached to the sugar are often refured to in reference to the carbon number they are bound to. For example, the phosphate residue is attached to the 5′ carbon of the sugar and the hydroxyl group is attached to the 3′ carbon of the sugar. We will often use the carbon number to refer to functional groups on nucleotides so be very familiar with the structure of the pentose sugar.
The pentose sugar in DNA is called deoxyribose, and in RNA, the sugar is ribose. The difference between the sugars is the presence of the hydroxyl group on the 2' carbon of the ribose and its absence on the 2' carbon of the deoxyribose. Hence you can determine if you are looking at a DNA or RNA nucleotide by the presence or absence of the hydroxyl group on the 2' carbon atom - you will likely be asked to do so on numerous occasions (including exams).
The Phosphate Group
There can be anywhere between 1 and 3 phosphate groups bound to the 5' carbon of the sugar. When one phosphate bound the nucleotide is referred to as a Nucleotide MonoPhosphate (NMP). If 2 phosphates are bound the nucleotide is referred to as Nucleotide DiPhosphate (NDP). When 3 phosphates are bound to the nucleotide it is referred to as a Nucleotide TriPhosphate (NTP). The phosphoanhydride bonds between that link the phosphate groups to each other have specific chemical properties that make them good for various biological functions. The hydrolysis of the bonds between the phosphate groups is thermodynamically exergonic in biological conditions and nature has evolved numerous mechanisms to couple this negative change in free-energy to help drive many reactions in the cell. The figure below shows an example of the hydrolysis of the nucleotide triphosphate ATP.
"High energy" bonds
The term "high energy bond" is used A LOT in biology. It is, however, one of those shortcuts we referred to earlier. The term refers to the amount of negative free energy associated with the HYDROLYSIS of that bond! The water is important. While we have tried to minimize the use of the vernacular "high energy" when referring to bonds, keep the above in mind when you are reading or listening to discussions in biology.
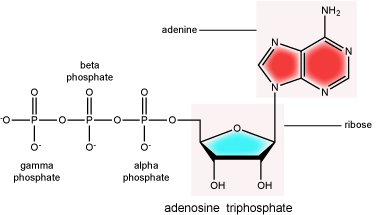
ATP (adenosine triphosphate) has three phosphate groups that can be removed by hydrolysis to form ADP (adenosine diphosphate) or AMP (adenosine monophosphate).The negative charges on the phosphate group naturally repel each other, requiring energy to bond them together and releasing energy when these bonds are broken.
Attribution: Marc T. Facciotti (original work)
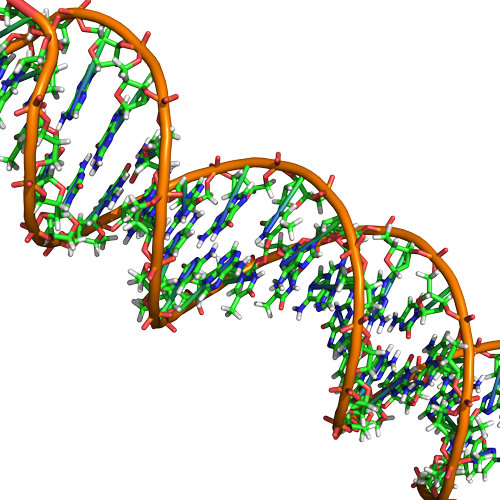
Double Helix Structure of DNA
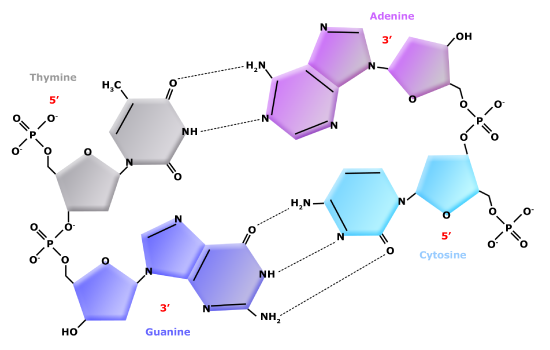
DNA has a double-helix structure (shown below). The sugar and phosphate lie on the outside of the helix, forming the backbone of the DNA. The nitrogenous bases are stacked in the interior, like the steps of a staircase, in pairs; the pairs are bound to each other by hydrogen bonds. Every base pair in the double helix is separated from the next base pair by 0.34 nm. The two strands of the helix run in opposite directions, meaning that the 5′ carbon end of one strand will face the 3′ carbon end of its matching strand. This is referred to as antiparallel orientation.
In a double helix, certain combinations of base pairing are chemically more favored than others based on the types and locations of functional groups on the nitrogenous bases of each nucleotide. In biology we find that adenine (A) is chemically complementary with thymidine (T) and guanine (G) is chemically complementary with cytosine (C), as shown below. We often refer to this pattern as "base complementarity" and say that the antiparallel strands are complementary to each other. For example, if the sequence of one strand is of DNA is 5'-AATTGGCC-3', the complementary strand would have the sequence 5'-GGCCAATT-3'.
Functions and roles of nucleic acids and nucleotides
Nucleic acids play a variety of roles in in cellular process besides being the information storage molecule. Nucleic acids, RNA in particular, are believed to be the first biologically active molecules during a period referred to as the "RNA world" when catalytic RNA were thought to serve the dual role as catalysts and information storing molecules. Remnants of the RNA world can be seen in many riboprotein complexes essential for life. In these RNA-Protein complexes the RNA serves both catalytic and structural roles. Examples include of such complexes include, ribosomes, RNases, splicesosome complexes and Telomerase. Nucleotides such as ATP and GTP also serve as mobile short-term energy transport units for the cell. Nucleotides also play important roles as co-factors (in addition to energy vehicles) for many enzymatic reactions. Like lipids, proteins and carbohydrates, nucleic acids and nucleotides play a wide variety of roles in the cell.
Genomes as organismal blueprints
A genome, not to be confused with a gnome, is an organism's complete collection of heritable information stored in DNA. - This means that: differences in information content help to explain the diversity of life we see all around us; - that changes to the information encoded in the genome are the primary drivers the phenotypic diversity we see (and some we can't) around us that are filtered by natural selection - thus the drivers of evolution Leads to questions: - if every cell in a multi-cellular organism contains the same sequence of DNA how can there be different cell types (e.g. how can a cell in a liver be so different from a cell in the brain if they both carry the same DNA)? - how do we read the information?
Determining a genome sequence
The information encoded in genomes provides important data for understanding life, its functions, its diversity and its evolution. It, therefore, stands to reason that a reasonable place to begin studies in biology would be to read the information content encoded in the genome(s) in question. A good starting point is to determine the sequence of nucleotides (A, G, C, T) and their organization into one or more independently replicating units of DNA (e.g. think chromosomes and/or plasmids ). For 30+ years after the discovery that DNA was the hereditary material this was a daunting proposition. In the late 1980's, however, the advent of semi-automated tools for DNA sequencing were pioneered and this began a revolution that has dramatically changed how we approach the study of life. Twenty years later in the mid-2000's, we entered a period of accelerated technological progress in which advances in materials sciences (particularly advances in our ability to make things on a very small scale), optics, electrical and computer engineering, bioengineering and computer sciences have all converged to bring us dramatic increases in our capacity to sequence DNA and correspondingly dramatic decreases in the cost to sequence numerous advances in our ability to sequence DNA. A famous example to illustrate this point is to compare the changes in cost to sequence the human genome. The first draft of the human genome took nearly 15 years and $3 billion dollars to complete. Today, 10's of human genomes can be sequenced in a single day on a single instrument at a cost of less than $1000 each (the cost and time continue to decrease). Today, companies like Illumina, Pacific Biosciences, Oxford Nanopore and others offer competing technologies that are driving down the cost and increasing the volume, quality, speed and portability of DNA sequencing.
One of the very exciting elements of the DNA sequencing revolution is that it has - and continues to - require contributions from biologists, chemists, materials scientists, electrical engineers, mechanical engineers, computer scientists and programmers, mathematicians and statisticians, product developers, and many other technical experts. The potential applications and implications of unlocking barriers to DNA sequencing has also engaged investors, business people, product developers, entrepreneurs, ethicists, policy makers, and many others in pursuit of new opportunities and in the need to think about how to best and most responsibly use this growing technology.
The technological advances in genome sequencing have resulted in a virtual flood of complete genome sequences being determined and deposited into publicly available databases. You can find many of them at the National Center for Biotechnology Information. The number of available completely sequenced genomes numbers in the tens of thousands - over 2000 eukaryotic genomes, over 600 archaeal genomes, and nearly 12000 bacterial genomes. Tens of thousands more genome sequencing projects are in progress. With this many genome sequences available - or soon to be available - we can start asking many questions about what we see in these genomes. What patterns are common to all genomes? How many genes are encoded in genomes? How are these organized? How many different types of features can we find? What do the features that we find do? How different are the genomes from one another? Is there evidence that can tell us how genomes evolve? Let's briefly examine a few of these quesitons.
Diversity of genomes
Diversity of sizes, number of genes, and chromosomes
Let's start by examining the range of genome sizes. In the table below we see a sampling of genomes from the database. We can see that the genomes of free living organism range tremendously in size. The smallest known genome is encoded in 580,000 base pairs while the largest is 150 billion base pairs - for reference, recall that the human genome is 3.2 billion base pairs. That's a huge range of sizes. Similar disparities in the number of genes also exist.
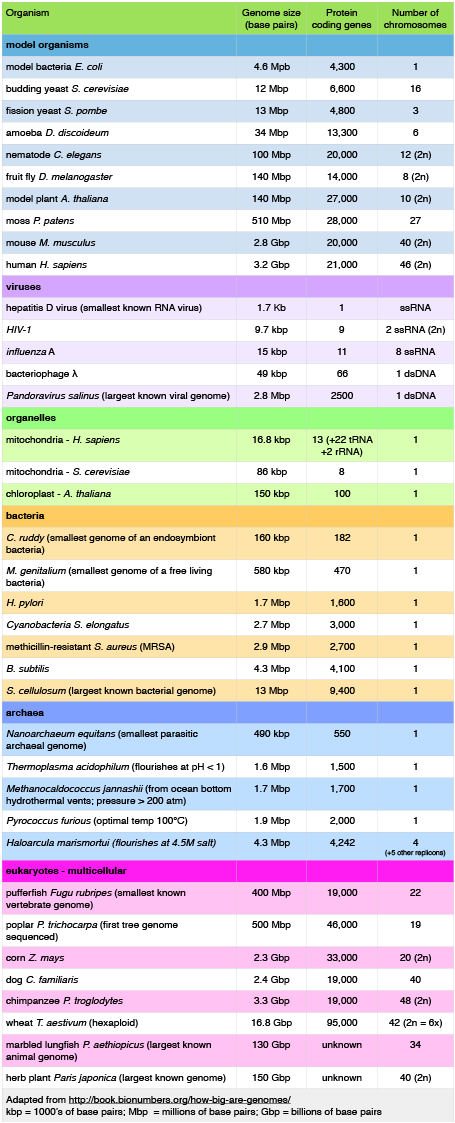
A sample of genome sizes, number of genes and chromosome copies. 2n = diploid number
Attribution: Marc T. Facciotti (own work - reproduced from http://book.bionumbers.org/how-big-are-genomes/)
Examining the table also reveals that some organisms carry with them more than one chromosome. Some genomes are also polyploid, meaning that they maintain multiple copies of similar but not identical (so called homologous) copies of each chromosome. A diploid organism carries in its genome 2 homologous copies (usually one from Mom and one from Dad) of each chromosome. Humans are diploid. Our somatic cells carry 2 homologous copies of 23 chromosomes. We received 23 copies of individual chromosomes from our mother and 23 copies from our father, for a total of 46. Some plants have higher ploidy. For example, a plant with four homologous copies of each chromosome is termed tetraploid. An organism with a single copy of each chromosome is termed haploid.
Structure of genomes
The table also provides clues to other points of interest. For instance, if we compare the pufferfish genome to the chimpanzee genome we note that they encode roughly the same number of genes (19,000) but they do so on dramatically differently sized genomes - 400 million base pairs versus 3.3 billion base pairs, respectively. That implies that the pufferfish genome must have much less space between its genes than what might be expected to be found in the chimpanzee genome. Indeed, this is the case and it the difference in gene density is not unique to these two genomes. If we look at the figure immediately below that attempts to represent a 50kb part of the human genome, we notice that in addition to the protein coding regions (indicated in red and pink) that many other so called "features" can be read from the genome. Many of these elements are contain highly repetitive sequences.
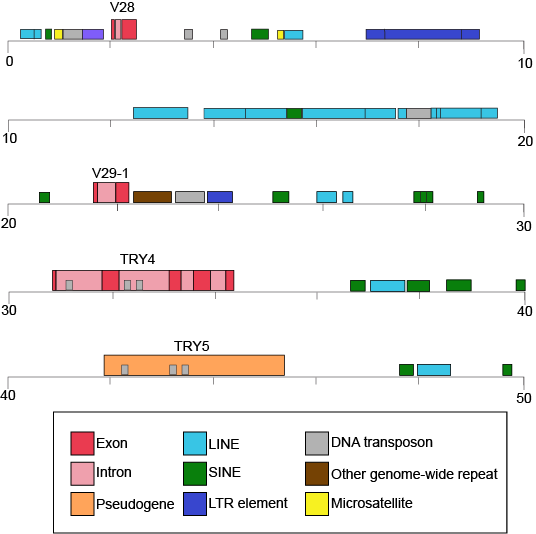
50-kb segment of the human β T-cell receptor locus on chromosome 7. This figure depicts a small region of the human genome and the types of "features" that can be read and decoded in the genome including but also in addition to protein coding sequences. Red and pink correspond to regions that encode proteins. Other colors represent different types of genomic elements. Attribution: Marc T. Facciotti (own work - reproduced from www.ncbi.nlm.nih.gov/books/NBK21134/)
If we now look at what fraction of the whole human genome each of these types of elements makes up - see the following figure - we see that protein coding genes only make up 48 million of the 3.2 billion bases of the haploid genome.
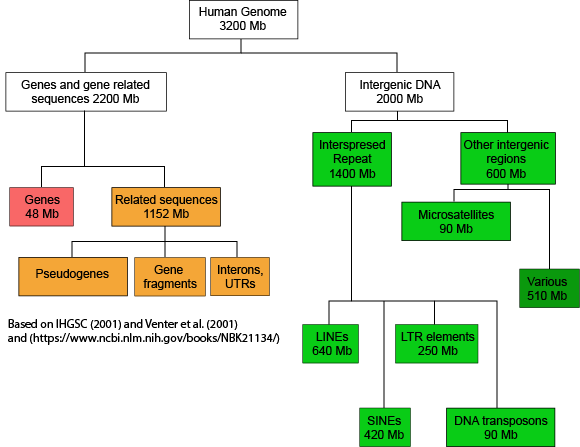
A graph depicting how the many base pairs of DNA in the human haploid genome are distributed between various identifiable features. Note only a small fraction of the genome is associated directly with protein coding regions. Attribution: Marc T. Facciotti (own work - reproduced from sources noted in figure)
When we examine the frequency of repeat regions versus protein coding regions in different species, we note large differences in protein coding versus non-coding regions.
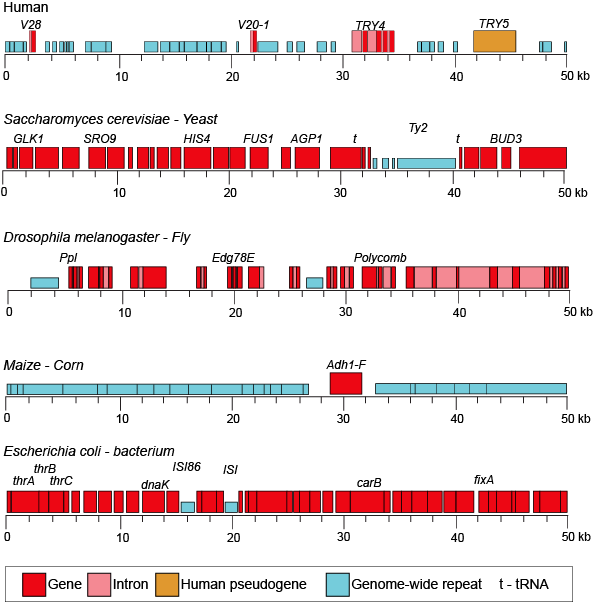
50-kb segments of different genomes illustrating the highly variable frequency of repeat versus protein coding elements in different species.
Attribution: Marc T. Facciotti (own work - reproduced from www.ncbi.nlm.nih.gov/books/NBK21134/)
Suggested discussion
Propose a hypothesis for why you think some genomes might have more or less non-coding sequences.
Dynamics of genome structure
Genomes change over time and numerous different types of events can change their sequence.
- Mutations - either accumulated during DNA replication or through environmental exposure to chemical mutagens or radiation. These changes typically occur at the level of single nucleotides.
- Genome rearrangements - this describes a class of large-scale changes that can occur and they include: (a) deletions - where segments of the chromosome are lost, (b) duplication - where regions of the chromosome are inadvertently duplicated, (c) insertions - the insertion of genetic material (sometimes this is acquired from viruses or the environment) - deletion/insertion pairs may happen across chromosomes, (d) inversions - where regions of the genome are flipped within the same chromosome and (e) translocations - where segments of the chromosome are translocated or moved elsewhere in the chromosome.
These changes happen at different rates and some are facilitated by the activity of enzyme of catalysts (e.g. transposases).
The study of genomes
Comparative genomics
One of the most common things to do with a collection of genome sequences is to compare the sequences of multiple genomes to one another. In general terms these types of activities fall under the umbrella of a field called comparative genomics.
Comparing the genomes of people who suffer from an inheritable disease to the genomes of people that are not afflicted can help us to uncover the genetic basis for the malady. Comparing the gene content, order and sequence of related microbes can help us find the genetic basis why some microbes cause disease while their close cousins are virtually harmless. We can compare genomes to understand how a new species may have evolved. There are many possible analyses! The basis of these analyses is similar: look for differences across multiple genomes and try to associate those difference with different traits or behaviors in those organisms.
Lastly, some people are comparing genome sequences to try to understand the evolutionary history of the organisms. Typically, these types of comparisons result in a graph known as a phylogenetic tree which is a graphical model of the evolutionary relationship between the various species being compared. This field, not surprisingly, is called phylogenomics.
Metagenomics: who is living somewhere and what are they doing?
In addition to studying the genomes of individual species the increasingly powerful DNA sequencing technologies are making it possible to simultaneously sequence the genomes of environmental samples that are inhabited by many different species. This field is called metagenomics. These studies are typically focused on trying to understand what microbial species inhabit different environments. There is great interest in using DNA sequencing to study the populations of microbes in the gut and to watch how the population changes in response different diets, to see if there is any association between the abundance of different microbes and various diseases, or to look for the presence of pathogens. People are using DNA sequencing of environmental metagenomic samples to explore which microbes inhabit different environments on Earth (from the deep sea, to soil, to air, to hyper-saline ponds, to cat feces, to some of the common surfaces we touch every day. If you can think of it, these days it seems as if someone has tried to sequence the microbes there.
In addition to discovering "who lives where", the sequencing of microbial populations in different environments can also reveal what protein-coding genes are present in an environment. This can give investigators clues into what metabolic activities might be occurring in that environment. In addition to providing important information about what kind of chemistry might be happening in a specific environment the catalog of genes that is accumulated can also serve as an important resource for the discovery of novel enzymes for applications in biotechnology.