Solutions for May 20, 2008, Second Midterm
- Page ID
- 6848
\[\mathrm{BioSci\: 102-02\\ May\: 20,\: 2008,\: Second\: Midterm}\]
Instructions:
• There are nine pages in this exam including the cover sheet, please count them before you start to make sure all are present.
• Write your name on each page of the exam.
• Write your answers in the space provided below each question. If you need more space use the back of the page and indicate clearly that you have continued your answer on the back. Do not use additional paper. For calculation problems a clear line of calculations leading to your final answer must be shown to obtain full credit.
Use the following \(\mathrm{pK_{a}}\) values for the exam.
| Amino Acid | \(\mathbf{\alpha -NH_{2}}\) | \(\mathbf{\alpha -COOH}\) | R-group |
|---|---|---|---|
| Ala, Gly, Ile, Leu, Val | 9.7 | 2.3 | \(-\) |
| Asn, Gln | 9.0 | 2.1 | \(-\) |
| Met, Ser, Trp | 9.3 | 2.3 | \(-\) |
| Phe | 9.1 | 1.8 | \(-\) |
| Pro | 10.6 | 2.0 | \(-\) |
| Thr | 10.4 | 2.6 | \(-\) |
| Asp | 9.8 | 2.1 | 3.9 |
| Glu | 9.7 | 2.2 | 4.2 |
| His | 9.2 | 1.8 | 6.0 |
| Cys | 10.8 | 1.8 | 8.3 |
| Tyr, Lys | 9.1 | 2.2 | 10.0 |
| Arg | 9.0 | 2.2 | 12.5 |
\(\begin{matrix} \mathrm{log\: 1.0=0.00}\\ \mathrm{log\: 1.5=0.18} \\ \mathrm{log\: 2.0=0.18}\\ \mathrm{log\: 2.5=0.40}\\ \mathrm{log\: 3.0=0.48}\\ \mathrm{log\: 3.5=0.54}\\ \mathrm{log\: 4.0=0.60}\\ \mathrm{log\: 4.5=0.65}\\ \mathrm{log\: 5.0=0.70}\\ \mathrm{log\: 5.5=0.74}\\ \mathrm{log\: 6.0=0.78}\\ \mathrm{log\: 6.5= 0.81}\\ \mathrm{log\: 7.0=0.85}\\ \mathrm{log\: 7.5=0.88}\\ \mathrm{log\: 8.0=0.90}\\ \mathrm{log\: 8.5=0.93}\\ \mathrm{log\: 9.0=0.95}\\ \mathrm{log\: 9.5=0.98}\\ \mathrm{log\: 10.0=1.00} \end{matrix}\)
\(\mathbf{R=8.3\times 10^{-3}\: kJ/ ^{\circ}K\cdot mol}\)
\(\mathbf{25^{\circ}C=298K}\)
| Question | Value | Score |
|---|---|---|
| 1 | 8 | |
| 2 | 21 | |
| 3 | 15 | |
| 4 | 12 | |
| 5 | 12 | |
| 6 | 16 | |
| 7 | 9 | |
| 8 | 14 | |
| 9 | 12 | |
| 10 | 10 | |
| 11 | 9 | |
| 12 | 12 | |
| Total | 150 |
1. The figure below represents an isoelectric focusing (IEF) gel on which three protein samples (A, B, and C) were run to completion. The locations of the two electrodes are indicated by the “+” and "-" signs.
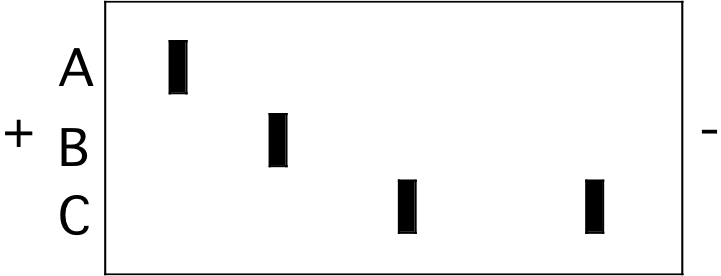
A B \(\mathrm{\fbox{C}}\) High pI is a basic protein that will initially have a “+” charge and will move toward the “-“ pole.
The illustration below shows the result of sodium dodecyl sulfate (SDS, and also including ß-mercaptoethanol) polyacrylamide gel electrophoresis of the same samples that were examined on the IEF gel above. The gel was stained with commassie blue dye. The direction of migration was from top to bottom.
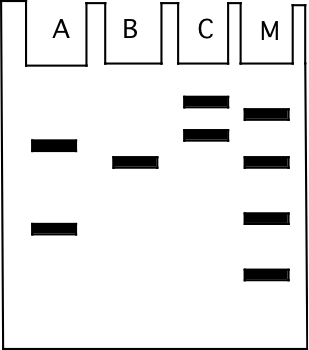
b) (4) The size markers used on the gel are in the lane labeled “M” and are 10 kilodaltons (kD), 20 kD, 50 kD, and 90 kD in molecular weight. What is the approximate molecular weight of the protein visible in sample “B”?
50 kD, smaller proteins move more rapidly through the gel so the markers go from smallest to largest going from bottom to top of the gel. The band in B migrates with the 50 kD marker.
2. In the space to the left of each of descriptions below, write the letters corresponding to all of the methods to which that statement applies. All correct answers must be given for full credit. (3 points each)
A. Gel filtration.
B. Isoelectric focusing gel electrophoresis.
C. Ion exchange chromatography.
D. Ligand affinity chromatography.
E. Antibody columns.
F. SDS polyacrylamide gel electrophoresis.
G. Reverse phase chromatography.
B, C can separate proteins on the basis of pI.
A, C, D, E, G can be readily adapted for purification of large amounts of protein.
D, E can commonly be used to purify a protein from a complex mixture in a single step.
A, F separates proteins on the basis of size/molecular weight.
G would likely work well to separate integral membrane proteins from soluble cytosolic proteins.
D requires some knowledge of the activity of the protein for separation.
F proteins are denatured when separated by this method.
3. (15) You have identified a new compound (call it "X") which, when added to a solution, significantly weakens salt bridges without affecting other types of weak chemical interactions. X is added to a tube of hemoglobin at pH7 at equilibrium with air and with a standard concentration of BPG. For each of the following, circle the letter to indicate if the addition of X will lead to an increase (I), decrease (D), or leave unchanged (U) the listed property of the hemoglobin solution (relative to the solution without added X). (3 points each). Numerous salt bridges stabilize the deoxy state. Weakening these salt bridges would favor the oxy state.
\(\mathbf{I\:\:\:\:\:D\:\:\:\:\:\fbox{U}}\) The fraction of hemoglobin that exhibits quaternary structure.
\(\mathbf{I\:\:\:\:\: \fbox{D}\:\:\:\:\:U}\) The amount of BPG bound by the hemoglobin.
\(\mathbf{\fbox{I}\:\:\:\:\:D\:\:\:\:\:U}\) The affinity of the hemoglobin for oxygen.
\(\mathbf{I\:\:\:\:\: \fbox{D}\:\:\:\:\:U}\) The amount of \(\mathrm{H^{+}}\) bound by the hemoglobin.
\(\mathbf{I\:\:\:\:\: \fbox{D}\:\:\:\:\:U}\) The \(\mathrm{P_{50}}\) of the hemoglobin for \(\mathrm{O_{2}}\).
4. Indicate which of the following statements about human hemoglobin (Hb) are true (T) or false (F) by circling the letter to the left of the statement. Assume that all solutions containing Hb also initially contain an equimolar concentration of 2,3-bisphosphoglycerate (BPG). (4 points each).
\(\mathrm{\fbox{T}\:\:\:\:F}\) If the \(\mathrm{P_{CO_{2}}}\) in a solution of Hb is decreased from 10 to 5 mm Hg, the oxygen saturation of the hemoglobin will increase. \(\mathbf{CO_{2}}\) promotes \(\mathbf{O_{2}}\) release
\(\mathrm{\fbox{T}\:\:\:\:F}\) An increase in the \(\mathrm{P_{CO_{2}}}\) in a tube containing Hb that is 40% saturated with \(\mathrm{O_{2}}\) will increase the fraction of Hb that is protonated. Higher \(\mathbf{P_{CO_{2}}}\) promotes the deoxy form, deoxy form binds more protons
\(\mathrm{T\:\:\:\: \fbox{F}}\) \(\mathrm{His_{146}(\beta)}\) participates in a salt bridge only in the deoxygenated form of adult Hb. An increase in \(\mathrm{P_{O_{2}}}\) over a solution of adult Hb initially at 30% saturation will lead to an increase in the \(\mathrm{pK_{a}}\) of this histidine. Higher \(\mathbf{P_{O_{2}}}\) means more oxy form, less salt bridge, less stabilization of protonated form, more \(\mathbf{H^{+}}\) release = lower \(\mathbf{pK_{a}}\)
5. The enzyme, “E”, catalyzes the reaction \(\mathrm{A\Leftrightarrow B+C}\). The \(\mathrm{\Delta G^{\circ’}}\) for the uncatalyzed reaction is +26.2 kJ/mol. Place an X next to the one correct answer to each of the following questions. (4 pts each).
a) The enzyme E will
always consume A faster than in the uncatalyzed reaction.
force the \(\mathrm{\Delta G}\) for the forward reaction to be negative.
make the \(\mathrm{\Delta G^{\circ’}}\) for the reaction less positive
X make B disappear faster than in an uncatalyzed reaction only if the concentration of B is greater than the equilibrium value.
change \(\mathrm{\Delta S}\) for the reaction, but not \(\mathrm{\Delta H}\).
b) Which is likely to bind more tightly to E?
A
C
X a structure intermediate between A and B + C
a combination of A, B, and C together
another enzyme
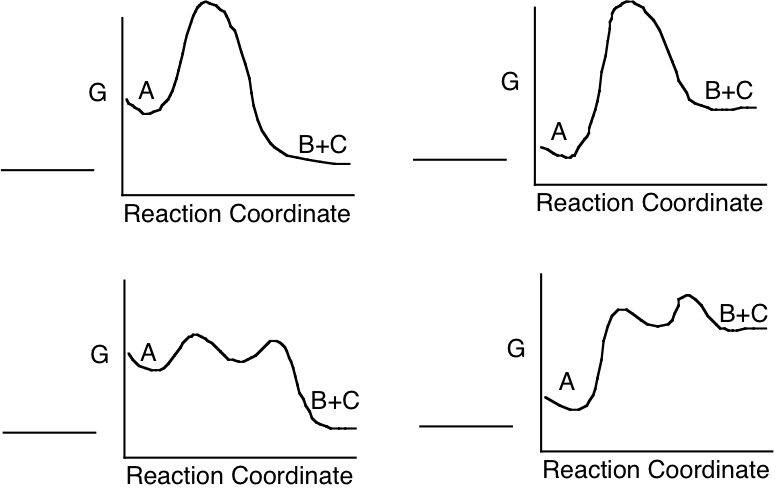
c) Which of the following energy diagrams is most likely to illustrate the progress of the enzyme catalyzed reaction in the direction as written, under standard state conditions?
6. (16) In the lungs of a particular mammal, the \(\mathrm{P_{O_{2}}}\) in the blood is 90 mm Hg and the pH is 7.6. In the tissues, the \(\mathrm{P_{O_{2}}}\) in the blood is 30 mm Hg and the pH is 7.0. The hemoglobin (Hb) of this mammal has a Hill coefficient (the exponent in the equation of Y versus \(\mathrm{P_{O_{2}}}\)) of 2.7. The \(\mathrm{P_{50}}\) for \(\mathrm{O_{2}}\) of this mammal’s Hb is 35 at pH 7.0 and 30 at pH 7.6. What fraction of the \(\mathrm{O_{2}}\) bound by the Hb of this mammal in the lungs is released to the tissues under these conditions (ignoring any effects of [\(\mathrm{CO_{2}}\)])?
\[\mathbf{Lungs-Y=\dfrac{P_{O_{2}}^{2.7}}{P_{O_{2}}^{2.7}+P_{50}^{2.7}}=\dfrac{90^{2.7}}{90^{2.7}+30^{2.7}}=0.951}\]
\[\mathbf{Tissues-Y=\dfrac{30^{2.7}}{30^{2.7}+35^{2.7}}=0.397}\]
so 0.951-0.397 = 0.554 of total oxygen capacity delivered.
Fraction of bound delivered = delivered/bound in lungs = 0.554/0.951 = 0.583 or 58.3%
7. (9) Below are alignments between human \(\alpha\)-globin corresponding regions of human myoglobin, zebrafish (a small fresh water fish) \(\alpha\)-globin, and horse \(\alpha\)-globin (not necessarily in that order). Between the alignments “|” indicates identical amino acids and “:” and “.” indicate very similar and largely similar amino acids, respectively. From the alignments, determine which sequence is which and write the letters corresponding to the sequences in the blanks below.

8. True-False: For each of the following statements circle "T" if the statement true and an "F" if the statement is false. (2 points each).
\(\mathbf{T\:\:\:\:\: \fbox{F}}\) Hydrophobic interactions between amino acid side chains (R-groups) are the most important factor in protein secondary structure. backbone interaction
\(\mathbf{\fbox{T}\:\:\:\:\:F}\) Hydrogen bonds between backbone carbonyl oxygen atoms and the hydrogen atoms of backbone NH groups are responsible for the stabilization of an \(\alpha\)-helix in a protein.
\(\mathbf{\fbox{T}\:\:\:\:\:F}\) All proteins with quaternary structure have more than one carboxyl terminus.
\(\mathbf{T\:\:\:\:\: \fbox{F}}\) Proteins that contain parallel \(\beta\)-pleated sheets do not contain \(\beta\)-turns.
\(\mathbf{T\:\:\:\:\: \fbox{F}}\) Cysteine and methionine are the two amino acids that commonly participate in the formation of disulfide bonds. Only Cys
\(\mathbf{T\:\:\:\:\: \fbox{F}}\) Chaperonins change the \(\Delta\)S of protein folding, but leave the \(\Delta\)H unchanged.
Don’t change either.
\(\mathbf{T\:\:\:\:\: \fbox{F}}\) Chaperonins are important for bringing proteins together to assemble into complexes because proteins are present at very low concentrations in cells.
(Proteins are present at high concentration and aggregation must be prevented)
9. For each of the biologically derived compounds below, fill in the letters corresponding to ALL of the properties which apply to that compound (3 pts each blank).
A. Net negative charge at pH 7.
B. Contains a cis double bond.
C. Contains a trans double bond.
D. Contains a carbon that is bound to both a nitrogen and an oxygen.
E. A glycolipid.
C,D, E A membrane sphingolipid that contains a palmitic acid residue and that has a single glucose residue attached directly to a hydroxyl group on the sphingosine backbone.
A A phosphatidic acid containing palmitic and stearic acid residues.
B A glycerolipid containing a palmitic, and two oleic acid residues.
A, B The fatty acid specified by the code \(20:2^{\Delta 12,15}\).
10. The structures below illustrate glyceraldehyde-3-phosphate bound to the active site of triose phosphate isomerase.
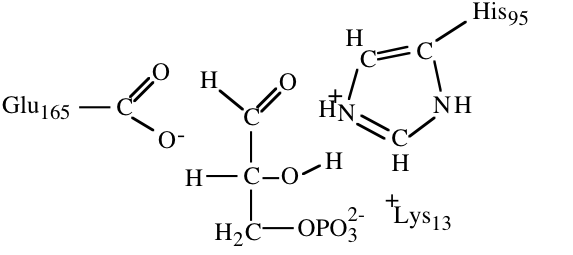
- a) (4) In an early step in the initial reaction of triose phosphate isomerase on glyceraldehydes-3-phosphate a part of the enzyme acts as a proton acceptor to facilitate catalysis. Circle the atom that accepts a proton in this step and draw a square around the proton that it accepts.
b) (6) The histidine at the active site acts as a proton acceptor in one part of the reaction mechanism. The unprotonated form of histidine is a much weaker base (weaker proton acceptor) than is the unprotonated form of lysine. If the histidine in triose phosphate isomerase were replaced with a lysine, would you expect the enzyme to become more or less active? Briefly explain your answer.
Less active. The different shape of the Lys would place it in the wrong location to accept a proton for optimal catalysis. In addition, the His acts as both a proton donor and acceptor in different steps of catalysis and the Lys would be a very weak donor.
11. Several different enzymes have been found that can cleave specific linkages in glycerophospholipids. The specificities of two of these enzymes are:
| Enzyme | Activity |
|---|---|
| Phospholipase A1 | Catalyzes hydrolysis of ester linkage between C1 of glycerol backbone and attached fatty acid side chain |
| Phospholipase D | Catalyzes hydrolysis of linkage between the phosphate and an attached additional moleucle (leaving the phosphate attached to the glycerol backbone). |
Two samples of phosphatidyl serine (with two oleic acids as fatty acid side chains) were separately digested with each of these different enzymes. Each digestion resulted in production of two new molecules, with one always being larger (higher in molecular weight) than the other.
Describe the type of structure (e. g. micelle, bilayer, lipid globule, liposome, dissolved in solution, etc.) that each of the following would form if it were mixed vigorously with water (assume pH 7). (3 pts each)
a) The small product of digestion with phospholipase D.
in solution, serine is the small product
b) The large product of digestion with phospholipase D.
liposomes, phophatidic acid is the large product
c) The small product of digestion with phospholipase A1.
micelles, oleic acid is the small product
12. You have isolated a protein from human liver and are studying it stability. On heating the protein you find that it first denatures at \(\mathrm{65^{\circ}C}\) (338K). You also measure the enthalpy of folding (\(\mathrm{\Delta H}\)) of this protein to be -85 kJ/mol.
a) (6) What is the entropy (\(\mathrm{\Delta S}\)) of folding of this protein?
\[\mathbf{at\: 338K\: \Delta G=0=\Delta H -T\Delta S}\]
\[\mathbf{ T\Delta S =\Delta H}\]
\[\mathbf{338 \Delta S=-85\: kJ/mol}\]
\[\mathbf{\Delta S=-85/338\: kJ/K\cdot mol=-0.25\: kJ/K\cdot mol}\]
\(\mathrm{\Delta S= \underline{-0.25\: kJ/K\cdot mol}}\)
(continued on following page) units
b) (6) If this protein were allowed to spontaneously fold in solution at \(\mathrm{25^{\circ}C}\), what effect would the folding reaction have on the temperature of the surrounding solution? (circle correct answer)
remain unchanged \(\mathrm{\fbox{increase}}\) decrease
Why?
The \(\mathrm{\Delta H}\) is negative so the reaction gives off heat. The heat will raise the temperature of the surrounding liquid.

