6.2.2: Regulating Transcription at the Chromatin Level
- Page ID
- 25751
↵
Learning Objectives
- Predict regions of the genome that are more likely to have “open” or “closed” chromatin and understand how acetylation affects chromatin.
- Identify the molecular nature of DNA methylation, the types of bases that are methylated, and the effect of DNA methylation on gene expression.
Eukaryotes regulate transcription via promoter sequences close to the transcription unit (as in prokaryotes) and also use more distant enhancer sequences to provide more variation in the timing, level, and location of transcription, however, there are still additional levels of genetic control. This consists of two major mechanism: (1) large-scale changes in chromatin structure, and (2) modification of bases in the DNA sequence. These two are often inter-connected.
Chromatin remodeling
Despite the simplified way in which we often represent DNA, DNA is almost always associated with various chromatin proteins. For example, histones remain associated with the DNA even during transcription. Thus the rate of transcription is also controlled by the accessibility of DNA to RNA polymerase and regulatory proteins. So, in regions with highly compact chromatin transcription is unlikely, even if all the necessary cis- and trans- factors are present in the nucleus.
The extent of chromatin compaction in various regions is regulated through the action of chromatin remodeling proteins. These protein complexes include enzymes that add or remove chemical tags, such as methyl or acetyl groups, to various DNA bound proteins, often the histone proteins in nucleosomes. These post-translational modifications alter the local chromatin density and thus the availability for transcription. Acetylated histones, for example, tend to be associated with actively transcribed genes, whereas deacetylated histone are associated with genes that are silenced.
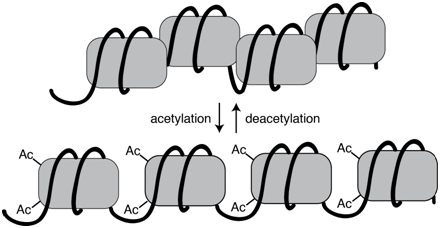
Figure \(\PageIndex{1}\): Acetylation of histone proteins is associated with more a more open chromatin configuration. Acetylation is a reversible process. (Origianl-Deyholos-CC:AN)
Likewise, methylation of DNA itself is also associated with transcription regulation. Cytosine bases, particularly when followed by a guanine are important targets for DNA methylation. Using a p to represent the phosphodiester bond between the nucleotides, these are often called CpG sites, and regions in which many CpG sites are nearby are known as CpG islands. Methylated cytosine within clusters of CpG sites is often associated with transcriptionally inactive DNA.
Thinking about CpG
Why is C followed by G an important site for DNA methylation? Think about the sequence of both strands of DNA. What is the complementary sequence of 5' ...CG... 3' ? Does this allow both strands to be methylated?
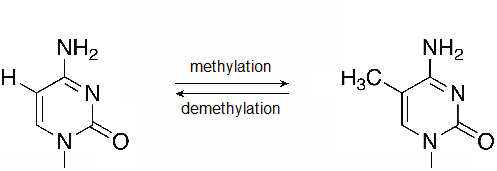
Figure \(\PageIndex{2}\): A methylation reaction produces 5-methylcytosine (5mC). Methyl groups may also be removed by DNA demethylases. (flickr-Beardy Git-CC:AND)
Common post-translational modifications to histones
These post-translational modifications are the result of enzymes that covalently link functional groups to amino acids at specific positions in histones.
- Acetylation
- Methylation
- Phosphorylation
- Ubiquitination
Combinations of these modifications was first postulated to be a "code" by which the regulation of genes is interpreted by David Allis and colleagues in 2001.
Heritability of Chromatin
The modification of DNA and its associated proteins is enzymatically reversible (acetylases/deacetylases; methyltranferases/demethylases) and thus a cyclical activity. However, when DNA is replicated, existing modifications on the parental strand will be added to the newly synthesized strand as well, maintaining the pattern in daughter cells until the modification is enzymatically removed. Regulation of these chemical modifications provides another layer through which eukaryotic cells control the transcription of specific genes.
Code readers
In addition to influencing an overall level of chromatin accessibility, modified histones may interact specifically with proteins that recognize their modifications to promote recruitment of additional proteins for functions in repair, replication, or transcription.
Contributors and Attributions
Dr. Todd Nickle and Isabelle Barrette-Ng (Mount Royal University) The content on this page is licensed under CC SA 3.0 licensing guidelines.


